3D genome tuner: compare multiple circular genomes in a 3D context
- PMID: 19944387
- PMCID: PMC5054402
- DOI: 10.1016/S1672-0229(08)60043-1
3D genome tuner: compare multiple circular genomes in a 3D context
Abstract
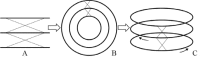
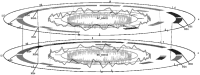
Circular genomes, being the largest proportion of sequenced genomes, play an important role in genome analysis. However, traditional 2D circular map only provides an overview and annotations of genome but does not offer feature-based comparison. For remedying these shortcomings, we developed 3D Genome Tuner, a hybrid of circular map and comparative map tools. Its capability of viewing comparisons between multiple circular maps in a 3D space offers great benefits to the study of comparative genomics. The program is freely available (under an LGPL licence) at http://sourceforge.net/projects/dgenometuner.
Figures
Similar articles
-
Taverna: a tool for building and running workflows of services.Nucleic Acids Res. 2006 Jul 1;34(Web Server issue):W729-32. doi: 10.1093/nar/gkl320. Nucleic Acids Res. 2006. PMID: 16845108 Free PMC article.
-
Genome Projector: zoomable genome map with multiple views.BMC Bioinformatics. 2009 Jan 23;10:31. doi: 10.1186/1471-2105-10-31. BMC Bioinformatics. 2009. PMID: 19166610 Free PMC article.
-
CMap3D: a 3D visualization tool for comparative genetic maps.Bioinformatics. 2010 Jan 15;26(2):273-4. doi: 10.1093/bioinformatics/btp646. Epub 2009 Nov 25. Bioinformatics. 2010. PMID: 19942584
-
Visualizing and comparing circular genomes using the CGView family of tools.Brief Bioinform. 2019 Jul 19;20(4):1576-1582. doi: 10.1093/bib/bbx081. Brief Bioinform. 2019. PMID: 28968859 Free PMC article. Review.
-
A brief introduction to web-based genome browsers.Brief Bioinform. 2013 Mar;14(2):131-43. doi: 10.1093/bib/bbs029. Epub 2012 Jul 3. Brief Bioinform. 2013. PMID: 22764121 Review.
Cited by
-
bioWeb3D: an online webGL 3D data visualisation tool.BMC Bioinformatics. 2013 Jun 7;14:185. doi: 10.1186/1471-2105-14-185. BMC Bioinformatics. 2013. PMID: 23758781 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

