Structure and functionality in flavivirus NS-proteins: perspectives for drug design
- PMID: 19945487
- PMCID: PMC3918146
- DOI: 10.1016/j.antiviral.2009.11.009
Structure and functionality in flavivirus NS-proteins: perspectives for drug design
Abstract
Flaviviridae are small enveloped viruses hosting a positive-sense single-stranded RNA genome. Besides yellow fever virus, a landmark case in the history of virology, members of the Flavivirus genus, such as West Nile virus and dengue virus, are increasingly gaining attention due to their re-emergence and incidence in different areas of the world. Additional environmental and demographic considerations suggest that novel or known flaviviruses will continue to emerge in the future. Nevertheless, up to few years ago flaviviruses were considered low interest candidates for drug design. At the start of the European Union VIZIER Project, in 2004, just two crystal structures of protein domains from the flaviviral replication machinery were known. Such pioneering studies, however, indicated the flaviviral replication complex as a promising target for the development of antiviral compounds. Here we review structural and functional aspects emerging from the characterization of two main components (NS3 and NS5 proteins) of the flavivirus replication complex. Most of the reviewed results were achieved within the European Union VIZIER Project, and cover topics that span from viral genomics to structural biology and inhibition mechanisms. The ultimate aim of the reported approaches is to shed light on the design and development of antiviral drug leads.
Copyright 2009 Elsevier B.V. All rights reserved.
Figures

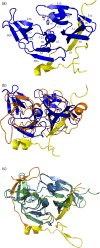
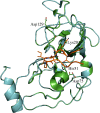
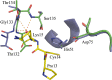
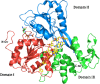
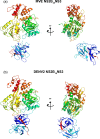

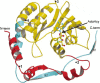
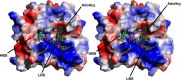

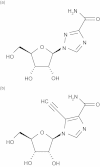
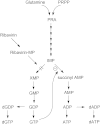
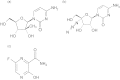
References
-
- Ackermann M., Padmanabhan R. De novo synthesis of RNA by the dengue virus RNA-dependent RNA polymerase exhibits temperature dependence at the initiation but not elongation phase. J. Biol. Chem. 2001;276:39926–39937. - PubMed
-
- Adachi T., Ago H., Habuka N., Okuda K., Komatsu M., Ikeda S., Yatsunami K. The essential role of C-terminal residues in regulating the activity of hepatitis C virus RNA-dependent RNA polymerase. Biochim. Biophys. Acta. 2002;1601:38–48. - PubMed
-
- Ago H., Adachi T., Yoshida A., Yamamoto M., Habuka N., Yatsunami K., Miyano M. Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus. Structure. 1999;7:1417–1426. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

