Visualizing protein-DNA interactions at the single-molecule level
- PMID: 19945909
- PMCID: PMC2819561
- DOI: 10.1016/j.cbpa.2009.10.035
Visualizing protein-DNA interactions at the single-molecule level
Abstract
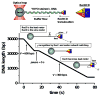
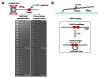
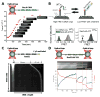
Recent advancements in single-molecule methods have allowed researchers to directly observe proteins acting on their DNA targets in real-time. Single-molecule imaging of protein-DNA interactions permits detection of the dynamic behavior of individual complexes that otherwise would be obscured in ensemble experiments. The kinetics of these processes can be monitored directly, permitting identification of unique subpopulations or novel reaction intermediates. Innovative techniques have been developed to isolate and manipulate individual DNA or protein molecules, and to visualize their interactions. The actions of proteins that have been visualized include: duplex DNA unwinding, DNA degradation, DNA packaging, translocation on DNA, sliding, superhelical twisting, and DNA bending, extension, and condensation. These single-molecule studies have provided new insights into nearly all aspects of DNA metabolism. Here we focus primarily on recent advances in fluorescence imaging and mechanical detection of individual protein-DNA complexes, with emphasis on selected proteins involved in DNA recombination: DNA helicases, DNA translocases, and DNA strand exchange proteins.
2009 Elsevier Ltd. All rights reserved.
Figures

References
-
- Zlatanova J, van Holde K. Single-molecule biology: what is it and how does it work? Mol Cell. 2006;24:317–329. - PubMed
-
- Cornish PV, Ha T. A survey of single-molecule techniques in chemical biology. ACS Chem Biol. 2007;2:53–61. - PubMed
-
- Park H, Toprak E, Selvin PR. Single-molecule fluorescence to study molecular motors. Q Rev Biophys. 2007;40:87–111. - PubMed
-
- Moffitt JR, Chemla YR, Smith SB, Bustamante C. Recent advances in optical tweezers. Annu Rev Biochem. 2008;77:205–228. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

