ABC transporters in Saccharomyces cerevisiae and their interactors: new technology advances the biology of the ABCC (MRP) subfamily
- PMID: 19946134
- PMCID: PMC2786581
- DOI: 10.1128/MMBR.00020-09
ABC transporters in Saccharomyces cerevisiae and their interactors: new technology advances the biology of the ABCC (MRP) subfamily
Abstract
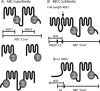
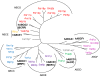
Members of the ATP-binding cassette (ABC) transporter superfamily exist in bacteria, fungi, plants, and animals and play key roles in the efflux of xenobiotic compounds, physiological substrates, and toxic intracellular metabolites. Based on sequence relatedness, mammalian ABC proteins have been divided into seven subfamilies, ABC subfamily A (ABCA) to ABCG. This review focuses on recent advances in our understanding of ABC transporters in the model organism Saccharomyces cerevisiae. We propose a revised unified nomenclature for the six yeast ABC subfamilies to reflect the current mammalian designations ABCA to ABCG. In addition, we specifically review the well-studied yeast ABCC subfamily (formerly designated the MRP/CFTR subfamily), which includes six members (Ycf1p, Bpt1p, Ybt1p/Bat1p, Nft1p, Vmr1p, and Yor1p). We focus on Ycf1p, the best-characterized yeast ABCC transporter. Ycf1p is located in the vacuolar membrane in yeast and functions in a manner analogous to that of the human multidrug resistance-related protein (MRP1, also called ABCC1), mediating the transport of glutathione-conjugated toxic compounds. We review what is known about Ycf1p substrates, trafficking, processing, posttranslational modifications, regulation, and interactors. Finally, we discuss a powerful new yeast two-hybrid technology called integrated membrane yeast two-hybrid (iMYTH) technology, which was designed to identify interactors of membrane proteins. iMYTH technology has successfully identified novel interactors of Ycf1p and promises to be an invaluable tool in future efforts to comprehensively define the yeast ABC interactome.
Figures





References
-
- Adamis, P. D. B., A. D. Panek, and E. C. A. Eleutherio. 2007. Vacuolar compartmentation of the cadmium-glutathione complex protects Saccharomyces cerevisiae from mutagenesis. Toxicol. Lett. 173:1-7. - PubMed
-
- Ambudkar, S. V., S. Dey, C. A. Hrycyna, M. Ramachandra, I. Pastan, and M. M. Gottesman. 1999. Biochemical, cellular, and pharmacological aspects of the multidrug transporter. Annu. Rev. Pharmacol. Toxicol. 39:361-398. - PubMed
-
- Bakos, E., R. Evers, G. Calenda, G. E. Tusnady, G. Szakacs, A. Varadi, and B. Sarkadi. 2000. Characterization of the amino-terminal regions in the human multidrug resistance protein (MRP1). J. Cell Sci. 113(Pt. 24):4451-4461. - PubMed
-
- Ballatori, N., C. L. Hammond, J. B. Cunningham, S. M. Krance, and R. Marchan. 2005. Molecular mechanisms of reduced glutathione transport: role of the MRP/CFTR/ABCC and OATP/SLC21A families of membrane proteins. Toxicol. Appl. Pharmacol. 204:238-255. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

