Pathogenomics of the virulence plasmids of Escherichia coli
- PMID: 19946140
- PMCID: PMC2786578
- DOI: 10.1128/MMBR.00015-09
Pathogenomics of the virulence plasmids of Escherichia coli
Erratum in
- Microbiol Mol Biol Rev. 2010 Sep;74(3):477-8
Abstract
Bacterial plasmids are self-replicating, extrachromosomal elements that are key agents of change in microbial populations. They promote the dissemination of a variety of traits, including virulence, enhanced fitness, resistance to antimicrobial agents, and metabolism of rare substances. Escherichia coli, perhaps the most studied of microorganisms, has been found to possess a variety of plasmid types. Included among these are plasmids associated with virulence. Several types of E. coli virulence plasmids exist, including those essential for the virulence of enterotoxigenic E. coli, enteroinvasive E. coli, enteropathogenic E. coli, enterohemorrhagic E. coli, enteroaggregative E. coli, and extraintestinal pathogenic E. coli. Despite their diversity, these plasmids belong to a few plasmid backbones that present themselves in a conserved and syntenic manner. Thanks to some recent research, including sequence analysis of several representative plasmid genomes and molecular pathogenesis studies, the evolution of these virulence plasmids and the implications of their acquisition by E. coli are now better understood and appreciated. Here, work involving each of the E. coli virulence plasmid types is summarized, with the available plasmid genomic sequences for several E. coli pathotypes being compared in an effort to understand the evolution of these plasmid types and define their core and accessory components.
Figures


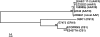



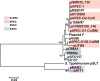
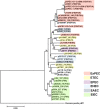
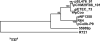
References
-
- Actis, L. A., M. E. Tolmasky, and J. H. Crosa. 1999. Bacterial plasmids: replication of extrachromosomal genetic elements encoding resistance to antimicrobial compounds. Front. Biosci. 4:D43-D62. - PubMed
-
- Aguero, M., G. de la Fuente, E. Vivaldi, and F. Cabello. 1989. ColV increases the virulence of Escherichia coli K1 strains in animal models of neonatal meningitis and urinary infection. Med. Microbiol. Immunol. 178:211-216. - PubMed
-
- An, H., J. M. Fairbrother, C. Desautels, and J. Harel. 1999. Distribution of a novel locus called paa (porcine attaching and effacing associated) among enteric Escherichia coli. Adv. Exp. Med. Biol. 473:179-184. - PubMed
-
- Ansuini, A., P. Candotti, G. Vecchi, V. Falbo, F. Minelli, and A. Caprioli. 1994. Necrotoxigenic E. coli in rabbits and horses. Vet. Res. 134:608. - PubMed
-
- Bakker, D., C. E. Vader, B. Roosendaal, F. R. Mooi, B. Oudega, and F. K. de Graaf. 1991. Structure and function of periplasmic chaperone-like proteins involved in the biosynthesis of K88 and K99 fimbriae in enterotoxigenic Escherichia coli. Mol. Microbiol. 5:875-886. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

