In vivo selection of tumor-targeting RNA motifs
- PMID: 19946274
- PMCID: PMC2795795
- DOI: 10.1038/nchembio.277
In vivo selection of tumor-targeting RNA motifs
Abstract
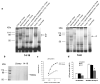
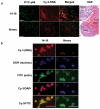
In an effort to target the in vivo context of tumor-specific moieties, we screened a large library of nuclease-resistant RNA oligonucleotides in tumor-bearing mice to identify candidate molecules with the ability to localize to hepatic colon cancer metastases. One of the selected molecules is an RNA aptamer that binds to p68, an RNA helicase that has been shown to be upregulated in colorectal cancer.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

