Crystal structure of prolyl 4-hydroxylase from Bacillus anthracis
- PMID: 19947658
- PMCID: PMC2806640
- DOI: 10.1021/bi901771z
Crystal structure of prolyl 4-hydroxylase from Bacillus anthracis
Abstract
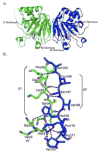
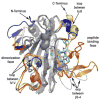
Prolyl 4-hydroxylases (P4H) catalyze the post-translational hydroxylation of proline residues and play a role in collagen production, hypoxia response, and cell wall development. P4Hs belong to the group of Fe(II)/alphaKG oxygenases and require Fe(II), alpha-ketoglutarate (alphaKG), and O(2) for activity. We report the 1.40 A structure of a P4H from Bacillus anthracis, the causative agent of anthrax, whose immunodominant exosporium protein BclA contains collagen-like repeat sequences. The structure reveals the double-stranded beta-helix core fold characteristic of Fe(II)/alphaKG oxygenases. This fold positions Fe-binding and alphaKG-binding residues in what is expected to be catalytically competent orientations and is consistent with proline peptide substrate binding at the active site mouth. Comparisons of the anthrax P4H structure with Cr P4H-1 structures reveal similarities in a peptide surface groove. However, sequence and structural comparisons suggest differences in conformation of adjacent loops may change the interaction with peptide substrates. These differences may be the basis of a substantial disparity between the K(M) values for the Cr P4H-1 compared to the anthrax and human P4H enzymes. Additionally, while previous structures of P4H enzymes are monomers, B. anthracis P4H forms an alpha(2) homodimer and suggests residues important for interactions between the alpha(2) subunits of alpha(2)beta(2) human collagen P4H. Thus, the anthrax P4H structure provides insight into the structure and function of the alpha-subunit of human P4H, which may aid in the development of selective inhibitors of the human P4H enzyme involved in fibrotic disease.
Figures




References
-
- Hieta R, Myllyharju J. Cloning and characterization of a low molecular weight prolyl 4-hydroxylase from arabidopsis thaliana. J Biol Chem. 2002;277:23965–23971. - PubMed
-
- Tiainen P, Myllyharju J, Koivunen P. Characterization of a second arabidopsis thaliana prolyl 4-hydroxylase with distinct substrate specificity. J Biol Chem. 2005;280:1142–1148. - PubMed
-
- Myllyharju J. Prolyl 4-hydroxylases, key enzymes in the synthesis of collagens and regulation of the response to hypoxia, and their roles as treatment targets. Ann Med Apr. 2008;23:1–16. - PubMed
-
- Myllyharju J, Kivirikko KI. Collagens, modifying enzymes and their mutations in humans, flies, and worms. Trends Genet. 2004;20:33–43. - PubMed
-
- Hieta R, Kukkola L, Permi P, Pirila P, Kivirikko KI, Kilpelainen I, Myllyharju J. The peptide-substrate-binding domain of human collagen prolyl 4-hydroxylases - backbone assignments, secondary structure, and binding of proline-rich peptides. J Biol Chem. 2003;278:34966–34974. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

