Eukaryotic transcriptomics in silico: optimizing cDNA-AFLP efficiency
- PMID: 19948029
- PMCID: PMC2797533
- DOI: 10.1186/1471-2164-10-565
Eukaryotic transcriptomics in silico: optimizing cDNA-AFLP efficiency
Abstract
Background: Complementary-DNA based amplified fragment length polymorphism (cDNA-AFLP) is a commonly used tool for assessing the genetic regulation of traits through the correlation of trait expression with cDNA expression profiles. In spite of the frequent application of this method, studies on the optimization of the cDNA-AFLP assay design are rare and have typically been taxonomically restricted. Here, we model cDNA-AFLPs on all 92 eukaryotic species for which cDNA pools are currently available, using all combinations of eight restriction enzymes standard in cDNA-AFLP screens.
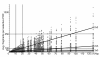
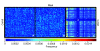
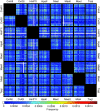
Results: In silco simulations reveal that cDNA pool coverage is largely determined by the choice of individual restriction enzymes and that, through the choice of optimal enzyme combinations, coverage can be increased from <40% to 75% without changing the underlying experimental design. We find evidence of phylogenetic signal in the coverage data, which is largely mediated by organismal GC content. There is nonetheless a high degree of consistency in cDNA pool coverage for particular enzyme combinations, indicating that our recommendations should be applicable to most eukaryotic systems. We also explore the relationship between the average observed fragment number per selective AFLP-PCR reaction and the size of the underlying cDNA pool, and show how AFLP experiments can be used to estimate the number of genes expressed in a target tissue.
Conclusion: The insights gained from in silico screening of cDNA-AFLPs from a broad sampling of eukaryotes provide a set of guidelines that should help to substantially increase the efficiency of future cDNA-AFLP experiments in eukaryotes. In silico simulations also suggest a novel use of cDNA-AFLP screens to determine the number of transcripts expressed in a target tissue, an application that should be invaluable as next-generation sequencing technologies are adapted for differential display.
Figures

References
-
- Breyne P, Dreesen R, Cannoot B, Rombaut D, Vandepoele K, Rombauts S, Vanderhaeghen R, Inze D, Zabeau M. Quantitative cDNA-AFLP analysis for genome-wide expression studies. Mol Genet Genomics. 2003;269:173–179. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

