Comparative study of human mitochondrial proteome reveals extensive protein subcellular relocalization after gene duplications
- PMID: 19948060
- PMCID: PMC2790464
- DOI: 10.1186/1471-2148-9-275
Comparative study of human mitochondrial proteome reveals extensive protein subcellular relocalization after gene duplications
Abstract
Background: Gene and genome duplication is the principle creative force in evolution. Recently, protein subcellular relocalization, or neolocalization was proposed as one of the mechanisms responsible for the retention of duplicated genes. This hypothesis received support from the analysis of yeast genomes, but has not been tested thoroughly on animal genomes. In order to evaluate the importance of subcellular relocalizations for retention of duplicated genes in animal genomes, we systematically analyzed nuclear encoded mitochondrial proteins in the human genome by reconstructing phylogenies of mitochondrial multigene families.
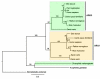
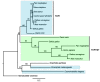
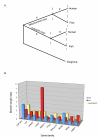
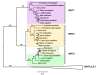
Results: The 456 human mitochondrial proteins selected for this study were clustered into 305 gene families including 92 multigene families. Among the multigene families, 59 (64%) consisted of both mitochondrial and cytosolic (non-mitochondrial) proteins (mt-cy families) while the remaining 33 (36%) were composed of mitochondrial proteins (mt-mt families). Phylogenetic analyses of mt-cy families revealed three different scenarios of their neolocalization following gene duplication: 1) relocalization from mitochondria to cytosol, 2) from cytosol to mitochondria and 3) multiple subcellular relocalizations. The neolocalizations were most commonly enabled by the gain or loss of N-terminal mitochondrial targeting signals. The majority of detected subcellular relocalization events occurred early in animal evolution, preceding the evolution of tetrapods. Mt-mt protein families showed a somewhat different pattern, where gene duplication occurred more evenly in time. However, for both types of protein families, most duplication events appear to roughly coincide with two rounds of genome duplications early in vertebrate evolution. Finally, we evaluated the effects of inaccurate and incomplete annotation of mitochondrial proteins and found that our conclusion of the importance of subcellular relocalization after gene duplication on the genomic scale was robust to potential gene misannotation.
Conclusion: Our results suggest that protein subcellular relocalization is an important mechanism for the retention and gain of function of duplicated genes in animal genome evolution.
Figures
Similar articles
-
Protein subcellular relocalization of duplicated genes in Arabidopsis.Genome Biol Evol. 2014 Sep 4;6(9):2501-15. doi: 10.1093/gbe/evu191. Genome Biol Evol. 2014. PMID: 25193306 Free PMC article.
-
Age distribution patterns of human gene families: divergent for Gene Ontology categories and concordant between different subcellular localizations.Mol Genet Genomics. 2014 Apr;289(2):137-47. doi: 10.1007/s00438-013-0799-8. Epub 2013 Dec 10. Mol Genet Genomics. 2014. PMID: 24322347
-
Genome duplication and gene-family evolution: the case of three OXPHOS gene families.Gene. 2008 Sep 15;421(1-2):1-6. doi: 10.1016/j.gene.2008.05.011. Epub 2008 Jun 23. Gene. 2008. PMID: 18573316 Review.
-
Whole genome duplications and expansion of the vertebrate GATA transcription factor gene family.BMC Evol Biol. 2009 Aug 20;9:207. doi: 10.1186/1471-2148-9-207. BMC Evol Biol. 2009. PMID: 19695090 Free PMC article.
-
Molluscan mitochondrial genomes break the rules.Philos Trans R Soc Lond B Biol Sci. 2021 May 24;376(1825):20200159. doi: 10.1098/rstb.2020.0159. Epub 2021 Apr 5. Philos Trans R Soc Lond B Biol Sci. 2021. PMID: 33813887 Free PMC article. Review.
Cited by
-
Mitochondrial Sco proteins are involved in oxidative stress defense.Redox Biol. 2019 Feb;21:101079. doi: 10.1016/j.redox.2018.101079. Epub 2018 Dec 12. Redox Biol. 2019. PMID: 30593977 Free PMC article.
-
Protein subcellular relocalization and function of duplicated flagellar calcium binding protein genes in honey bee trypanosomatid parasite.PLoS Genet. 2024 Mar 4;20(3):e1011195. doi: 10.1371/journal.pgen.1011195. eCollection 2024 Mar. PLoS Genet. 2024. PMID: 38437202 Free PMC article.
-
Recurrent sequence evolution after independent gene duplication.BMC Evol Biol. 2020 Aug 8;20(1):98. doi: 10.1186/s12862-020-01660-1. BMC Evol Biol. 2020. PMID: 32770961 Free PMC article.
-
Decoding the Divergent Subcellular Location of Two Highly Similar Paralogous LEA Proteins.Int J Mol Sci. 2018 May 31;19(6):1620. doi: 10.3390/ijms19061620. Int J Mol Sci. 2018. PMID: 29857468 Free PMC article.
-
Subcellular Relocalization and Positive Selection Play Key Roles in the Retention of Duplicate Genes of Populus Class III Peroxidase Family.Plant Cell. 2014 Jun;26(6):2404-2419. doi: 10.1105/tpc.114.124750. Epub 2014 Jun 16. Plant Cell. 2014. PMID: 24934172 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

