Autism susceptibility candidate 2 (Auts2) encodes a nuclear protein expressed in developing brain regions implicated in autism neuropathology
- PMID: 19948250
- PMCID: PMC2818569
- DOI: 10.1016/j.gep.2009.11.005
Autism susceptibility candidate 2 (Auts2) encodes a nuclear protein expressed in developing brain regions implicated in autism neuropathology
Abstract
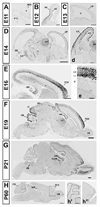
Autism susceptibility candidate 2 (Auts2) is a gene associated with autism and mental retardation, whose function is unknown. Expression of Auts2 mRNA and protein were studied in the developing mouse brain by in situ hybridization, immunohistochemistry, and western blotting. Auts2 mRNA was highly expressed in the developing cerebral cortex and cerebellum, regions often affected by neuropathological changes in autism, and a few other brain regions. On embryonic day (E) 12, Auts2 mRNA was expressed in the cortical preplate, where it colocalized with Tbr1, a transcription factor specific for postmitotic projection neurons. From E16 to postnatal day 21, Auts2 was expressed most abundantly in frontal cortex, hippocampus and cerebellum, including Purkinje cells and deep nuclei. High levels of Auts2 were also detected in developing dorsal thalamus, olfactory bulb, inferior colliculus and substantia nigra. Auts2 protein showed similar regional expression patterns as the mRNA. At the cellular level, Auts2 protein was localized in the nuclei of neurons and some neuronal progenitors.
Figures



References
-
- Amaral D, Schumann C, Nordahl C. Neuroanatomy of autism. Trends. Neurosci. 2008;31:137–145. - PubMed
-
- Arlotta P, Molyneaux BJ, Chen J, Inoue J, Kominami R, Macklis JD. Neuronal subtype-specific genes that control corticospinal motor neuron development in vivo. Neuron. 2005;45:207–221. - PubMed
-
- Bauman ML, Kemper TL. Neuroanatomic observations of the brain in autism: a review and future directions. Int. J. Dev. Neurosci. 2005;23(2–3):183–187. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

