Mammalian Rif1 contributes to replication stress survival and homology-directed repair
- PMID: 19948482
- PMCID: PMC2779251
- DOI: 10.1083/jcb.200902039
Mammalian Rif1 contributes to replication stress survival and homology-directed repair
Abstract
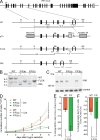
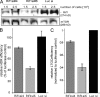
Rif1, originally recognized for its role at telomeres in budding yeast, has been implicated in a wide variety of cellular processes in mammals, including pluripotency of stem cells, response to double-strand breaks, and breast cancer development. As the molecular function of Rif1 is not known, we examined the consequences of Rif1 deficiency in mouse cells. Rif1 deficiency leads to failure in embryonic development, and conditional deletion of Rif1 from mouse embryo fibroblasts affects S-phase progression, rendering cells hypersensitive to replication poisons. Rif1 deficiency does not alter the activation of the DNA replication checkpoint but rather affects the execution of repair. RNA interference to human Rif1 decreases the efficiency of homology-directed repair (HDR), and Rif1 deficiency results in aberrant aggregates of the HDR factor Rad51. Consistent with a role in S-phase progression, Rif1 accumulates at stalled replication forks, preferentially around pericentromeric heterochromatin. Collectively, these findings reveal a function for Rif1 in the repair of stalled forks by facilitating HDR.
Figures


References
-
- Adams M.M., Wang B., Xia Z., Morales J.C., Lu X., Donehower L.A., Bochar D.A., Elledge S.J., Carpenter P.B. 2005. 53BP1 oligomerization is independent of its methylation by PRMT1. Cell Cycle. 4:1854–1861 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

