A dual function for Pex3p in peroxisome formation and inheritance
- PMID: 19948495
- PMCID: PMC2779223
- DOI: 10.1083/jcb.200906161
A dual function for Pex3p in peroxisome formation and inheritance
Abstract
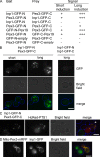
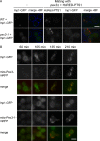
Saccharomyces cerevisiae Pex3p has been shown to act at the ER during de novo peroxisome formation. However, its steady state is at the peroxisomal membrane, where its role is debated. Here we show that Pex3p has a dual function: one in peroxisome formation and one in peroxisome segregation. We show that the peroxisome retention factor Inp1p interacts physically with Pex3p in vitro and in vivo, and split-GFP analysis shows that the site of interaction is the peroxisomal membrane. Furthermore, we have generated PEX3 alleles that support peroxisome formation but fail to support recruitment of Inp1p to peroxisomes, and as a consequence are affected in peroxisome segregation. We conclude that Pex3p functions as an anchor for Inp1p at the peroxisomal membrane, and that this function is independent of its role at the ER in peroxisome biogenesis.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

