Adenovirus RID-alpha activates an autonomous cholesterol regulatory mechanism that rescues defects linked to Niemann-Pick disease type C
- PMID: 19948501
- PMCID: PMC2779231
- DOI: 10.1083/jcb.200903039
Adenovirus RID-alpha activates an autonomous cholesterol regulatory mechanism that rescues defects linked to Niemann-Pick disease type C
Abstract
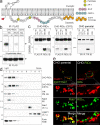
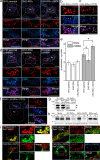
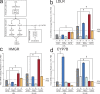
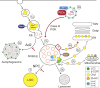
Host-pathogen interactions are important model systems for understanding fundamental cell biological processes. In this study, we describe a cholesterol-trafficking pathway induced by the adenovirus membrane protein RID-alpha that also subverts the cellular autophagy pathway during early stages of an acute infection. A palmitoylation-defective RID-alpha mutant deregulates cholesterol homeostasis and elicits lysosomal storage abnormalities similar to mutations associated with Niemann-Pick type C (NPC) disease. Wild-type RID-alpha rescues lipid-sorting defects in cells from patients with this disease by a mechanism involving a class III phosphatidylinositol-3-kinase. In contrast to NPC disease gene products that are localized to late endosomes/lysosomes, RID-alpha induces the accumulation of autophagy-like vesicles with a unique molecular composition. Ectopic RID-alpha regulates intracellular cholesterol trafficking at two distinct levels: the egress from endosomes and transport to the endoplasmic reticulum necessary for homeostatic gene regulation. However, RID-alpha also induces a novel cellular phenotype, suggesting that it activates an autonomous cholesterol regulatory mechanism distinct from NPC disease gene products.
Figures




References
-
- Biederbick A., Kern H.F., Elsässer H.P. 1995. Monodansylcadaverine (MDC) is a specific in vivo marker for autophagic vacuoles. Eur. J. Cell Biol. 66:3–14 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

