Calcium binds to LipL32, a lipoprotein from pathogenic Leptospira, and modulates fibronectin binding
- PMID: 19948735
- PMCID: PMC2823465
- DOI: 10.1074/jbc.M109.006320
Calcium binds to LipL32, a lipoprotein from pathogenic Leptospira, and modulates fibronectin binding
Abstract
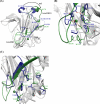
Tubulointerstitial nephritis is a cardinal renal manifestation of leptospirosis. LipL32, a major lipoprotein and a virulence factor, locates on the outer membrane of the pathogen Leptospira. It evades immune response by recognizing and adhering to extracellular matrix components of the host cell. The crystal structure of Ca(2+)-bound LipL32 was determined at 2.3 A resolution. LipL32 has a novel polyD sequence of seven aspartates that forms a continuous acidic surface patch for Ca(2+) binding. A significant conformational change was observed for the Ca(2+)-bound form of LipL32. Calcium binding to LipL32 was determined by isothermal titration calorimetry. The binding of fibronectin to LipL32 was observed by Stains-all CD and enzyme-linked immunosorbent assay experiments. The interaction between LipL32 and fibronectin might be associated with Ca(2+) binding. Based on the crystal structure of Ca(2+)-bound LipL32 and the Stains-all results, fibronectin probably binds near the polyD region on LipL32. Ca(2+) binding to LipL32 might be important for Leptospira to interact with the extracellular matrix of the host cell.
Figures



References
-
- Farr R. W. (1995) Clin. Infect. Dis. 21, 1–6 - PubMed
-
- Biegel E., Mortensen H. (1995) Ugeskr. Laeger 157, 153–157 - PubMed
-
- Bharti A. R., Nally J. E., Ricaldi J. N., Matthias M. A., Diaz M. M., Lovett M. A., Levett P. N., Gilman R. H., Willig M. R., Gotuzzo E., Vinetz J. M. (2003) Lancet 3, 757–771 - PubMed
-
- Yang C. W. (2007) Kidney Int. 72, 918–925 - PubMed
-
- Dolhnikoff M., Mauad T., Bethlem E. P., Carvalho C. R. (2007) Braz. J. Infect. Dis. 11, 142–148 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Miscellaneous

