Aminopeptidase fingerprints, an integrated approach for identification of good substrates and optimal inhibitors
- PMID: 19948737
- PMCID: PMC2823418
- DOI: 10.1074/jbc.M109.060418
Aminopeptidase fingerprints, an integrated approach for identification of good substrates and optimal inhibitors
Abstract
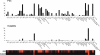
Aminopeptidases process the N-terminal amino acids of target substrates by sequential cleavage of one residue at a time. They are found in all cell compartments of prokaryotes and eukaryotes, being implicated in the major proteolytic events of cell survival, defense, growth, and development. We present a new approach for the fast and reliable evaluation of the substrate specificity of individual aminopeptidases. Using solid phase chemistry with the 7-amino-4-carbamoylmethylcoumarin fluorophore, we have synthesized a library of 61 individual natural and unnatural amino acids substrates, chosen to cover a broad spectrum of the possible interactions in the S1 pocket of this type of protease. As proof of concept, we determined the substrate specificity of human, pig, and rat orthologs of aminopeptidase N (CD13), a highly conserved cell surface protease that inactivates enkephalins and other bioactive peptides. Our data reveal a large and hydrophobic character for the S1 pocket of aminopeptidase N that is conserved with aminopeptidase Ns. Our approach, which can be applied in principle to all aminopeptidases, yields useful information for the design of specific inhibitors, and more importantly, reveals a relationship between the kinetics of substrate hydrolysis and the kinetics of enzyme inhibition.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

