Quantitative proteomic analysis of purified yeast kinetochores identifies a PP1 regulatory subunit
- PMID: 19948764
- PMCID: PMC2800092
- DOI: 10.1101/gad.1865909
Quantitative proteomic analysis of purified yeast kinetochores identifies a PP1 regulatory subunit
Erratum in
-
Corrigendum: Quantitative proteomic analysis of purified yeast kinetochores identifies a PP1 regulatory subunit.Genes Dev. 2016 Oct 15;30(20):2339. doi: 10.1101/gad.291450.116. Genes Dev. 2016. PMID: 27898395 Free PMC article. No abstract available.
Abstract
The kinetochore is a macromolecular complex that controls chromosome segregation and cell cycle progression. When sister kinetochores make bioriented attachments to microtubules from opposite poles, the spindle checkpoint is silenced. Biorientation and the spindle checkpoint are regulated by a balance between the Ipl1/Aurora B protein kinase and the opposing activity of protein phosphatase I (PP1). However, little is known about the regulation of PP1 localization and activity at the kinetochore. Here, we developed a method to purify centromere-bound kinetochores and used quantitative proteomics to identify the Fin1 protein as a PP1 regulatory subunit. The Fin1/PP1 complex is regulated by phosphorylation and 14-3-3 protein binding. When Fin1 is mislocalized, bipolar spindles fail to assemble but the spindle checkpoint is inappropriately silenced due to PP1 activity. These data suggest that Fin1 is a PP1 regulatory subunit whose spatial and temporal activity must be precisely controlled to ensure genomic stability.
Figures
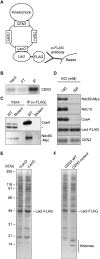


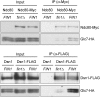



Comment in
-
Overcoming inhibition in the spindle checkpoint.Genes Dev. 2009 Dec 15;23(24):2799-805. doi: 10.1101/gad.1882109. Genes Dev. 2009. PMID: 20008930 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
