Thermodynamic analysis of ligand binding and ligand binding-induced tertiary structure formation by the thiamine pyrophosphate riboswitch
- PMID: 19948769
- PMCID: PMC2802028
- DOI: 10.1261/rna.1847310
Thermodynamic analysis of ligand binding and ligand binding-induced tertiary structure formation by the thiamine pyrophosphate riboswitch
Abstract
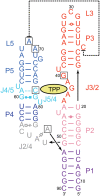
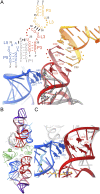
The thi-box riboswitch regulates gene expression in response to the intracellular concentration of thiamine pyrophosphate (TPP) in archaea, bacteria, and eukarya. To complement previous biochemical, genetic, and structural studies of this phylogenetically widespread RNA domain, we have characterized its interaction with TPP by isothermal titration calorimetry. This shows that TPP binding is highly dependent on Mg(2+) concentration. The dissociation constant decreases from approximately 200 nM at 0.5 mM Mg(2+) concentration to approximately 9 nM at 2.5 mM Mg(2+) concentration. Binding is enthalpically driven, but the unfavorable entropy of binding decreases as Mg(2+) concentration rises, suggesting that divalent cations serve to pre-organize the RNA. Mutagenesis, biochemical analysis, and a new crystal structure of the riboswitch suggest that a critical element that participates in organizing the riboswitch structure is the tertiary interaction formed between the P3 and L5 regions. This tertiary contact is distant from the TPP binding site, but calorimetric analysis reveals that even subtle mutations in L5 can have readily detectable effects on TPP binding. The thermodynamic signatures of these mutations, namely decreased favorable enthalpy of binding and small effects on entropy of binding, are consistent with the P3-L5 association contributing allosterically to TPP-induced compaction of the RNA.
Figures


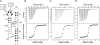
References
-
- Alam S, Grum-Tokars V, Krucinska J, Kundracik ML, Wedekind JE. Conformational heterogeneity at position U37 of an all-RNA hairpin ribozyme with implications for metal binding and the catalytic structure of the S-turn. Biochemistry. 2005;44:14396–14408. - PubMed
-
- Batey RT, Gilbert SD, Montange RK. Structure of a natural guanine-responsive riboswitch complexed with the metabolite hypoxanthine. Nature. 2004;432:411–415. - PubMed
-
- Brünger AT, Adams PD, Clore GM, Gros P, Grosse-Kunstleve RW, Jiang J-S, Kuszewski J, Nilges M, Pannu NS, Read RJ, et al. Crystallography and NMR system: A new software system for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. - PubMed
-
- Canny MD, Jucker FM, Kellogg E, Khvorova A, Jayasena SD, Pardi A. Fast cleavage kinetics of a natural hammerhead ribozyme. J Am Chem Soc. 2004;126:10848–10849. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
