Global regulation of gene expression and cell differentiation in Caulobacter crescentus in response to nutrient availability
- PMID: 19948804
- PMCID: PMC2812448
- DOI: 10.1128/JB.01240-09
Global regulation of gene expression and cell differentiation in Caulobacter crescentus in response to nutrient availability
Abstract
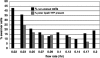
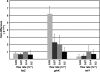
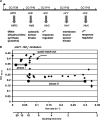
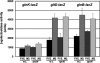
In a developmental strategy designed to efficiently exploit and colonize sparse oligotrophic environments, Caulobacter crescentus cells divide asymmetrically, yielding a motile swarmer cell and a sessile stalked cell. After a relatively fixed time period under typical culture conditions, the swarmer cell differentiates into a replicative stalked cell. Since differentiation into the stalked cell type is irreversible, it is likely that environmental factors such as the availability of essential nutrients would influence the timing of the decision to abandon motility and adopt a sessile lifestyle. We measured two different parameters in nutrient-limited chemostat cultures, biomass concentration and the ratio of nonstalked to stalked cells, over a range of flow rates and found that nitrogen limitation significantly extended the swarmer cell life span. The transcriptional profiling experiments described here generate the first comprehensive picture of the global regulatory strategies used by an oligotroph when confronted with an environment where key macronutrients are sparse. The pattern of regulated gene expression in nitrogen- and carbon-limited cells shares some features in common with most copiotrophic organisms, but critical differences suggest that Caulobacter, and perhaps other oligotrophs, have evolved regulatory strategies to deal distinctly with their natural environments. We hypothesize that nitrogen limitation extends the swarmer cell lifetime by delaying the onset of a sequence of differentiation events, which when initiated by the correct combination of external environmental cues, sets the swarmer cell on a path to differentiate into a stalked cell within a fixed time period.
Figures


References
-
- Ausmees, N., and C. Jacobs-Wagner. 2003. Spatial and temporal control of differentiation and cell cycle progression in Caulobacter crescentus. Annu. Rev. Microbiol. 57:225-247. - PubMed
-
- Biondi, E. G., S. J. Reisinger, J. M. Skerker, M. Arif, B. S. Perchuk, K. R. Ryan, and M. T. Laub. 2006. Regulation of the bacterial cell cycle by an integrated genetic circuit. Nature 444:899-904. - PubMed
-
- Biondi, E. G., J. M. Skerker, M. Arif, M. S. Prasol, B. S. Perchuk, and M. T. Laub. 2006. A phosphorelay system controls stalk biogenesis during cell cycle progression in Caulobacter crescentus. Mol. Microbiol. 59:386-401. - PubMed
-
- Blauwkamp, T. A., and A. J. Ninfa. 2003. Antagonism of PII signaling by the AmtB protein of Escherichia coli. Mol. Microbiol. 48:1017-1028. - PubMed
-
- Blauwkamp, T. A., and A. J. Ninfa. 2002. Physiological role of the GlnK signal transduction protein of Escherichia coli: survival of nitrogen starvation. Mol. Microbiol. 46:203-214. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

