Distribution of cepacian biosynthesis genes among environmental and clinical Burkholderia strains and role of cepacian exopolysaccharide in resistance to stress conditions
- PMID: 19948863
- PMCID: PMC2805225
- DOI: 10.1128/AEM.01828-09
Distribution of cepacian biosynthesis genes among environmental and clinical Burkholderia strains and role of cepacian exopolysaccharide in resistance to stress conditions
Abstract
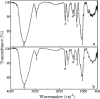
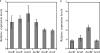
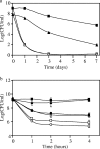
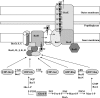
The genus Burkholderia includes strains pathogenic to animals and plants, bioremediators, or plant growth promoters. Genome sequence analyses of representative Burkholderia cepacia complex (Bcc) and non-Bcc strains for the presence of the bce-I gene cluster, directing the biosynthesis of the exopolysaccharide (EPS) cepacian, further extended this previously described cluster by another 9 genes. The genes in the bce-II cluster were named bceM to bceU and encode products putatively involved in nucleotide sugar precursor biosynthesis and repeat unit assembly, modification, and translocation across the cytoplasmic membrane. Disruption of the B. cepacia IST408 bceQ and bceR genes, encoding a putative repeat unit flippase and a glycosyltransferase, respectively, resulted in the abolishment of cepacian biosynthesis. A mutation in the bceS gene, encoding a putative acyltransferase, did not affect EPS production yield significantly but decreased its acetylation content by approximately 20%. Quantitative real-time reverse transcription-PCR experiments confirmed the induction of genes in the bce-I and bce-II clusters in a Burkholderia multivorans EPS producer clinical isolate in comparison to the level for its isogenic EPS-defective strain. Fourier Transform infrared spectroscopy analysis confirmed that the exopolysaccharide produced by 10 Burkholderia isolates tested was cepacian. The ability of Burkholderia strains to withstand desiccation and metal ion stress was higher when bacteria were incubated in the presence of 2.5 g/liter of cepacian, suggesting that this EPS plays a role in the survival of these bacteria by contributing to their ability to thrive in different environments.
Figures
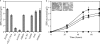
References
-
- Bartholdson, S. J., A. R. Brown, B. R. Mewburn, D. J. Clarke, S. C. Fry, D. J. Campopiano, and J. R. Govan. 2008. Plant host and sugar alcohol induced exopolysaccharide biosynthesis in the Burkholderia cepacia complex. Microbiology 154:2513-2521. - PubMed
-
- Buendia, A. M., B. Enenkel, R. Koplin, K. Niehaus, W. Arnold, and A. Puhler. 1991. The Rhizobium meliloti exoZl exoB fragment of megaplasmid 2: ExoB functions as a UDP-glucose 4-epimerase and ExoZ shows homology to NodX of Rhizobium leguminosarum biovar viciae strain TOM. Mol. Microbiol. 5:1519-1530. - PubMed
-
- Bullock, W. C., J. M. Fernandz, and J. M. Short. 1987. XL1-Blue: a high efficiency plasmid transforming recA Escherichia coli strain with beta-galactosidase selection. Biotechniques 5:376-379.
-
- Bylund, J., L. A. Burgess, P. Cescutti, R. K. Ernst, and D. P. Speert. 2006. Exopolysaccharides from Burkholderia cenocepacia inhibit neutrophil chemotaxis and scavenge reactive oxygen species. J. Biol. Chem. 281:2526-2532. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

