Quimp3, an automated pseudopod-tracking algorithm
- PMID: 19949291
- PMCID: PMC2852557
- DOI: 10.4161/cam.4.1.9953
Quimp3, an automated pseudopod-tracking algorithm
Abstract
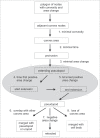
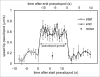
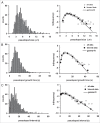
To understand movement of amoeboid cells we have developed an information tool that automatically detects protrusions of moving cells. The algorithm uses digitized cell recordings at a speed of approximately 1 image per second that are analyzed in three steps. In the first part, the outline of a cell is defined as a polygon of approximately 150 nodes, using the previously published Quimp2 program. By comparing the position of the nodes in place and time, each node contains information on position, local curvature and speed of movement. The second part uses rules for curvature and movement to define the position and time of start and end of a growing pseudopod. This part of the algorithm produces quantitative data on size, surface area, lifetime, frequency and direction of pseudopod extension. The third part of the algorithm assigns qualitative properties to each pseudopod. It decides on the origin of a pseudopod as splitting of an existing pseudopod or as extension de novo. It also decides on the fate of each pseudopod as merged with the cell body or retracted. Here we describe the pseudopod tool and present the first data based on the analysis of approximately 1,000 pseudopodia extended by Dictyostelium cells in the absence of external cues.
Figures


References
-
- Le Clainche C, Carlier MF. Regulation of actin assembly associated with protrusion and adhesion in cell migration. Physiol Rev. 2008;88:489–513. - PubMed
-
- Van Haastert PJM, Devreotes PN. Chemotaxis: signalling the way forward. Nat Rev Mol Cell Biol. 2004;5:626–634. - PubMed
-
- Pollard TD. Regulation of actin filament assembly by Arp2/3 complex and formins. Annu Rev Biophys Biomol Struct. 2007;36:451–477. - PubMed
-
- Arrieumerlou C, Meyer T. A local coupling model and compass parameter for eukaryotic chemotaxis. Dev Cell. 2005;8:215–227. - PubMed
-
- Costantino S, Kent CB, Godin AG, Kennedy TE, Wiseman PW, Fournier AE. Semi-automated quantification of filopodial dynamics. J Neurosci Methods. 2008;171:165–173. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
