Mouse models of genomic syndromes as tools for understanding the basis of complex traits: an example with the smith-magenis and the potocki-lupski syndromes
- PMID: 19949547
- PMCID: PMC2709937
- DOI: 10.2174/138920209788488508
Mouse models of genomic syndromes as tools for understanding the basis of complex traits: an example with the smith-magenis and the potocki-lupski syndromes
Abstract
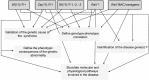
Each human's genome is distinguished by extra and missing DNA that can be "benign" or powerfully impact everything from development to disease. In the case of genomic disorders DNA rearrangements, such as deletions or duplications, correlate with a clinical specific phenotype. The clinical presentations of genomic disorders were thought to result from altered gene copy number of physically linked dosage sensitive genes. Genomic disorders are frequent diseases (~1 per 1,000 births). Smith-Magenis syndrome (SMS) and Potocki-Lupski syndrome (PTLS) are genomic disorders, associated with a deletion and a duplication, of 3.7 Mb respectively, within chromosome 17 band p11.2. This region includes 23 genes. Both syndromes have complex and distinctive phenotypes including multiple congenital and neurobehavioral abnormalities. Human chromosome 17p11.2 is syntenic to the 32-34 cM region of murine chromosome 11. The number and order of the genes are highly conserved. In this review, we will exemplify how genomic disorders can be modeled in mice and the advantages that such models can give in the study of genomic disorders in particular and gene copy number variation (CNV) in general. The contributions of the SMS and PTLS animal models in several aspects ranging from more specific ones, as the definition of the clinical aspects of the human clinical spectrum, the identification of dosage sensitive genes related to the human syndromes, to the more general contributions as the definition of genetic locus impacting obesity and behavior and the elucidation of general mechanisms related to the pathogenesis of gene CNV are discussed.
Keywords: Gene copy number variation; complex traits; mouse models.; phenotypic consequences.
Figures


References
-
- Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, Qi Y, Scherer SW, Lee C. Detection of large-scale variation in the human genome. Nat. Genet. 2004;36:949–951. - PubMed
-
- Sebat J, Lakshmi B, Troge J, Alexander J, Young J, Lundin P, Maner S, Massa H, Walker M, Chi M, Navin N, Lucito R, Healy J, Hicks J, Ye K, Reiner A, Gilliam TC, Trask B, Patterson N, Zetterberg A, Wigler M. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. - PubMed
-
- Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, An-drews TD, Fiegler H, Shapero MH, Carson AR, Chen W, Cho EK, Dallaire S, Freeman JL, Gonzalez JR, Gratacos M, Huang J, Kalaitzopoulos D, Komura D, Macdonald JR, Marshall CR, Mei R, Montgomery L, Nishimura K, Okamura K, Shen F, Somerville MJ, Tchinda J, Valsesia A, Woodwark C, Yang F, Zhang J, Zerjal T, Zhang J, Armen-gol L, Conrad DF, Estivill X, Tyler-Smith C, Carter NP, Aburatani H, Lee C, Jones KW, Scherer SW, Hurles ME. Global variation in copy number in the human genome. Nature. 2006;444:444–454. - PMC - PubMed
-
- Cheng Z, Ventura M, She X, Khaitovich P, Graves T, Osoe-gawa K, Church D, DeJong P, Wilson RK, Paabo S, Rocchi M, Eichler EE. A genome-wide comparison of recent chimpanzee and human segmental duplications. Nature. 2005;437:88–93. - PubMed
-
- Johnson ME, Viggiano L, Bailey JA, Abdul-Rauf M, Good-win G, Rocchi M, Eichler EE. Positive selection of a gene family during the emergence of humans and African apes. Nature. 2001;413:514–519. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
