Kinetic and structural characterization of dihydrofolate reductase from Streptococcus pneumoniae
- PMID: 19950924
- PMCID: PMC3773979
- DOI: 10.1021/bi901614m
Kinetic and structural characterization of dihydrofolate reductase from Streptococcus pneumoniae
Abstract
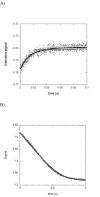
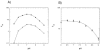
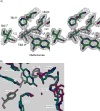
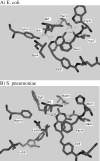
Drug resistance associated with dihydrofolate reductase (DHFR) has emerged as a critical issue in the treatment of bacterial infections. In our efforts to understand the mechanism of a drug-resistant dihydrofolate reductase (DHFR) from a pathogenic bacterial source, we report the first kinetic characterization of Streptococcus pneumoniae DHFR (spDHFR) along with its X-ray structure. This study revealed that the kinetic properties of spDHFR were significantly different from those of Escherichia coli DHFR. The product (tetrahydrofolate) dissociation step that is the rate-limiting step in E. coli DHFR is significantly accelerated in spDHFR so that hydride transfer or a preceding step is rate-limiting. Comparison of the binding parameters of this enzyme to those of a mutant spDHFR (Sp9) confirmed that the Leu100 residue in spDHFR is the critical element for the trimethoprim (TMP) resistance. Steady-state kinetics exhibited a pH dependence in k(cat), which prompted us to elucidate the role of the new catalytic residue (His33) in the active site of spDHFR. Structural data of the Sp9 mutant in complex with NADPH and methotrexate confirmed the participation of His33 in a hydrogen bonding network involving a water molecule, the hydroxyl group of Thr119, and the carboxylate ion of Glu30. Sequence analysis of the DHFR superfamily revealed that the His residue is the major amino acid component at this position and is found mostly in pathogenic bacterial DHFRs. A mutation of Val100 to Leu demonstrated a steric clash of the leucine side chain with the side chains of Ile8 and Phe34, rationalizing weaker binding of trimethoprim to Leu100 DHFR. Understanding the role of specific amino acids in the active site coupled with detailed structural analysis will inform us on how to better design inhibitors targeting drug-resistant pathogenic bacterial DHFRs.
Figures





Similar articles
-
Evidence for a functional role of the dynamics of glycine-121 of Escherichia coli dihydrofolate reductase obtained from kinetic analysis of a site-directed mutant.Biochemistry. 1997 Dec 16;36(50):15792-800. doi: 10.1021/bi9716231. Biochemistry. 1997. PMID: 9398309
-
Multiple mutations modulate the function of dihydrofolate reductase in trimethoprim-resistant Streptococcus pneumoniae.Antimicrob Agents Chemother. 2001 Apr;45(4):1104-8. doi: 10.1128/AAC.45.4.1104-1108.2001. Antimicrob Agents Chemother. 2001. PMID: 11257022 Free PMC article.
-
Crystal structures of wild-type and mutant methicillin-resistant Staphylococcus aureus dihydrofolate reductase reveal an alternate conformation of NADPH that may be linked to trimethoprim resistance.J Mol Biol. 2009 Apr 17;387(5):1298-308. doi: 10.1016/j.jmb.2009.02.045. Epub 2009 Feb 26. J Mol Biol. 2009. PMID: 19249312 Free PMC article.
-
Distal Regions Regulate Dihydrofolate Reductase-Ligand Interactions.Methods Mol Biol. 2021;2253:185-219. doi: 10.1007/978-1-0716-1154-8_12. Methods Mol Biol. 2021. PMID: 33315225 Review.
-
Multiple intermediates, diverse conformations, and cooperative conformational changes underlie the catalytic hydride transfer reaction of dihydrofolate reductase.Top Curr Chem. 2013;337:165-87. doi: 10.1007/128_2012_408. Top Curr Chem. 2013. PMID: 23420416 Free PMC article. Review.
Cited by
-
eModel-BDB: a database of comparative structure models of drug-target interactions from the Binding Database.Gigascience. 2018 Aug 1;7(8):giy091. doi: 10.1093/gigascience/giy091. Gigascience. 2018. PMID: 30052959 Free PMC article.
-
Temperature-Dependent Kinetic Isotope Effects in R67 Dihydrofolate Reductase from Path-Integral Simulations.J Phys Chem B. 2021 Feb 11;125(5):1369-1377. doi: 10.1021/acs.jpcb.0c10318. Epub 2021 Feb 1. J Phys Chem B. 2021. PMID: 33522797 Free PMC article.
-
Kinetic and chemical mechanism of the dihydrofolate reductase from Mycobacterium tuberculosis.Biochemistry. 2011 Jan 25;50(3):367-75. doi: 10.1021/bi1016843. Epub 2010 Dec 29. Biochemistry. 2011. PMID: 21138249 Free PMC article.
-
Crystal structure of dihydrofolate reductase from the filarial nematode W. bancrofti in complex with NADPH and folate.PLoS Negl Trop Dis. 2023 Apr 27;17(4):e0011303. doi: 10.1371/journal.pntd.0011303. eCollection 2023 Apr. PLoS Negl Trop Dis. 2023. PMID: 37104530 Free PMC article.
-
Simultaneous Control of Endogenous and User-Defined Genetic Pathways Using Unique ecDHFR Pharmacological Chaperones.Cell Chem Biol. 2020 May 21;27(5):622-634.e6. doi: 10.1016/j.chembiol.2020.03.006. Epub 2020 Apr 23. Cell Chem Biol. 2020. PMID: 32330442 Free PMC article.
References
-
- Hawser S, Lociuro S, Islam K. Dihydrofolate reductase inhibitors as antibacterial agents. Biochem Pharmacol. 2006;71:941–948. - PubMed
-
- Pikis A, Donkersloot JA, Rodriguez WJ, Keith JM. A conservative amino acid mutation in the chromosome-encoded dihydrofolate reductase confers trimethoprim resistance in Streptococcus pneumoniae. J Infect Dis. 1998;178:700–706. - PubMed
-
- Zhanel GG, Wang X, Nichol K, Nikulin A, Wierzbowski AK, Mulvey M, Hoban DJ. Molecular characterisation of Canadian paediatric multidrug-resistant Streptococcus pneumoniae from 1998–2004. Int J Antimicrob Agents. 2006;28:465–471. - PubMed
-
- Laue H, Weiss L, Bernardi A, Hawser S, Lociuro S, Islam K. In vitro activity of the novel diaminopyrimidine, iclaprim, in combination with folate inhibitors and other antimicrobials with different mechanisms of action. J Antimicrob Chemother. 2007;60:1391–1394. - PubMed
-
- Frey KM, Liu J, Lombardo MN, Bolstad DB, Wright DL, Anderson AC. Crystal structures of wild-type and mutant methicillinresistant Staphylococcus aureus dihydrofolate reductase reveal an alternate conformation of NADPH that may be linked to trimethoprim resistance. J Mol Biol. 2009;387:1298–1308. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

