Analysis of recent segmental duplications in the bovine genome
- PMID: 19951423
- PMCID: PMC2796684
- DOI: 10.1186/1471-2164-10-571
Analysis of recent segmental duplications in the bovine genome
Abstract
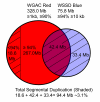
Background: Duplicated sequences are an important source of gene innovation and structural variation within mammalian genomes. We performed the first systematic and genome-wide analysis of segmental duplications in the modern domesticated cattle (Bos taurus). Using two distinct computational analyses, we estimated that 3.1% (94.4 Mb) of the bovine genome consists of recently duplicated sequences (>or= 1 kb in length, >or= 90% sequence identity). Similar to other mammalian draft assemblies, almost half (47% of 94.4 Mb) of these sequences have not been assigned to cattle chromosomes.
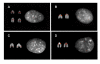
Results: In this study, we provide the first experimental validation large duplications and briefly compared their distribution on two independent bovine genome assemblies using fluorescent in situ hybridization (FISH). Our analyses suggest that the (75-90%) of segmental duplications are organized into local tandem duplication clusters. Along with rodents and carnivores, these results now confidently establish tandem duplications as the most likely mammalian archetypical organization, in contrast to humans and great ape species which show a preponderance of interspersed duplications. A cross-species survey of duplicated genes and gene families indicated that duplication, positive selection and gene conversion have shaped primates, rodents, carnivores and ruminants to different degrees for their speciation and adaptation. We identified that bovine segmental duplications corresponding to genes are significantly enriched for specific biological functions such as immunity, digestion, lactation and reproduction.
Conclusion: Our results suggest that in most mammalian lineages segmental duplications are organized in a tandem configuration. Segmental duplications remain problematic for genome and assembly and we highlight genic regions that require higher quality sequence characterization. This study provides insights into mammalian genome evolution and generates a valuable resource for cattle genomics research.
Figures


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

