Characterization of [4Fe-4S]-containing and cluster-free forms of Streptomyces WhiD
- PMID: 19954209
- PMCID: PMC2815329
- DOI: 10.1021/bi901498v
Characterization of [4Fe-4S]-containing and cluster-free forms of Streptomyces WhiD
Abstract
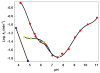
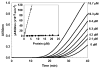
WhiD, a member of the WhiB-like (Wbl) family of iron-sulfur proteins found exclusively within the actinomycetes, is required for the late stages of sporulation in Streptomyces coelicolor. Like all other Wbl proteins, WhiD has not so far been purified in a soluble form that contains a significant amount of cluster, and characterization has relied on cluster-reconstituted protein. Thus, a major goal in Wbl research is to obtain and characterize native protein containing iron-sulfur clusters. Here we report the analysis of S. coelicolor WhiD purified anaerobically from Escherichia coli as a soluble protein containing a single [4Fe-4S](2+) cluster ligated by four cysteines. Upon exposure to oxygen, spectral features associated with the [4Fe-4S] cluster were lost in a slow reaction that unusually yielded apo-WhiD directly without significant concentrations of cluster intermediates. This process was found to be highly pH dependent with an optimal stability observed between pH 7.0 and pH 8.0. Low molecular weight thiols, including a mycothiol analogue and thioredoxin, exerted a small but significant protective effect against WhiD cluster loss, an activity that could be of physiological importance. [4Fe-4S](2+) WhiD was found to react much more rapidly with superoxide than with either oxygen or hydrogen peroxide, which may also be of physiological significance. Loss of the [4Fe-4S] cluster to form apoprotein destabilized the protein fold significantly but did not lead to complete unfolding. Finally, apo-WhiD exhibited negligible activity in an insulin-based disulfide reductase assay, demonstrating that it does not function as a general protein disulfide reductase.
Figures




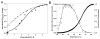

References
-
- den Hengst CD, Buttner MJ. Redox control in actinobacteria. Biochim Biophys Acta. 2008;1780:1201–1216. - PubMed
-
- Soliveri JA, Gomez J, Bishai WR, Chater KF. Multiple paralogous genes related to the Streptomyces coelicolor developmental regulatory gene whiB are present in Streptomyces and other actinomycetes. Microbiology. 2000;146(Pt 2):333–343. - PubMed
-
- Davis NK, Chater KF. The Streptomyces coelicolor whiB gene encodes a small transcription factor-like protein dispensable for growth but essential for sporulation. Mol Gen Genet. 1992;232:351–358. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

