Rapid turnover of mitochondrial uncoupling protein 3
- PMID: 19954423
- PMCID: PMC3661275
- DOI: 10.1042/BJ20091321
Rapid turnover of mitochondrial uncoupling protein 3
Abstract
UCP3 (uncoupling protein 3) and its homologues UCP2 and UCP1 are regulators of mitochondrial function. UCP2 is known to have a short half-life of approx. 1 h, owing to its rapid degradation by the cytosolic 26S proteasome, whereas UCP1 is turned over much more slowly by mitochondrial autophagy. In the present study we investigate whether UCP3 also has a short half-life, and whether the proteasome is involved in UCP3 degradation. UCP3 half-life was examined in the mouse C2C12 myoblast cell line by inhibiting protein synthesis with cycloheximide and monitoring UCP3 protein levels by immunoblot analysis. We show that UCP3 has a short half-life of 0.5-4 h. Rapid degradation was prevented by a cocktail of proteasome inhibitors, supporting a proteasomal mechanism for turnover. In addition, this phenotype is recapitulated in vitro: UCP3 was degraded in mitochondria isolated from rat skeletal muscle or brown adipose tissue with a half-life of 0.5-4 h, but only in the presence of a purified 26S proteasomal fraction. This in vitro proteolysis was also sensitive to proteasome inhibition. This phenotype is in direct contrast with the related proteins UCP1 and the adenine nucleotide translocase, which have long half-lives. Therefore UCP3 is turned over rapidly in multiple cell types in a proteasome-dependent manner.
Figures
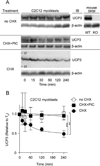
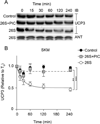

References
-
- Nicholls DG, Locke RM. Thermogenic mechanisms in brown fat. Physiol. Rev. 1984;64:1–64. - PubMed
-
- Cannon B, Nedergaard J. Brown adipose tissue: function and physiological significance. Physiol. Rev. 2004;84:277–359. - PubMed
-
- Pecqueur C, Alves-Guerra MC, Gelly C, Lévi-Meyrueis C, Couplan E, Collins S, Ricquier D, Bouillaud F, Miroux B. Uncoupling protein 2, in vivo distribution, induction upon oxidative stress, and evidence for translational regulation. J. Biol. Chem. 2001;276:8705–8712. - PubMed
-
- Brand MD, Esteves TC. Physiological functions of the mitochondrial uncoupling proteins UCP2 and UCP3. Cell Metab. 2005;2:85–93. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

