RNA profiles of rat olfactory epithelia: individual and age related variations
- PMID: 19954510
- PMCID: PMC2797534
- DOI: 10.1186/1471-2164-10-572
RNA profiles of rat olfactory epithelia: individual and age related variations
Abstract
Background: Mammalian genomes contain a large number (approximately 1000) of olfactory receptor (OR) genes, many of which (20 to 50%) are pseudogenes. OR gene transcription is not restricted to the olfactory epithelium, but is found in numerous tissues. Using microarray hybridization and RTqPCR, we analyzed the mRNA profiles of the olfactory epithelium of male and female Brown Norway rats of different origins and ages (newborn, adult and old).
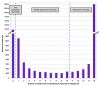
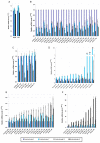
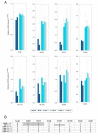
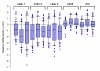
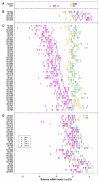
Results: (1) We observed very little difference between males and females and between rats from two different suppliers. (2) Different OR genes were expressed at varying levels, rather than uniformly across the four endoturbinates. (3) A large proportion of the gene transcripts (2/3 of all probes) were detected in all three age groups. Adult and older rats expressed similar numbers of OR genes, both expressing more OR genes than newborns. (4) Comparisons of whole transcriptomes or transcription profiles of expressed OR genes only showed a clear clustering of the samples as a function of age. (5) Most OR genes were expressed at lower levels at birth than in older animals, but a small number of OR genes were expressed specifically or were overexpressed in newborns.
Conclusion: Not all OR genes are expressed at a detectable level. Pups expressed fewer OR genes than adult rats, and generally at a lower level; however, a small subset of OR genes were more strongly expressed in these newborn rats. The reasons for these differences are not understood. However, the specific expression of some OR genes in newborn olfactory epithelia may be related to the blindness and deafness of pups at birth, when these pups are heavily reliant on olfaction and their mother.
Figures

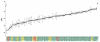



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

