Increasing phylogenetic resolution at low taxonomic levels using massively parallel sequencing of chloroplast genomes
- PMID: 19954512
- PMCID: PMC2793254
- DOI: 10.1186/1741-7007-7-84
Increasing phylogenetic resolution at low taxonomic levels using massively parallel sequencing of chloroplast genomes
Abstract
Background: Molecular evolutionary studies share the common goal of elucidating historical relationships, and the common challenge of adequately sampling taxa and characters. Particularly at low taxonomic levels, recent divergence, rapid radiations, and conservative genome evolution yield limited sequence variation, and dense taxon sampling is often desirable. Recent advances in massively parallel sequencing make it possible to rapidly obtain large amounts of sequence data, and multiplexing makes extensive sampling of megabase sequences feasible. Is it possible to efficiently apply massively parallel sequencing to increase phylogenetic resolution at low taxonomic levels?
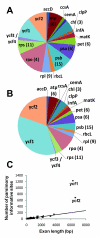
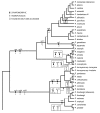
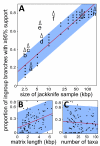
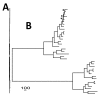
Results: We reconstruct the infrageneric phylogeny of Pinus from 37 nearly-complete chloroplast genomes (average 109 kilobases each of an approximately 120 kilobase genome) generated using multiplexed massively parallel sequencing. 30/33 ingroup nodes resolved with > or = 95% bootstrap support; this is a substantial improvement relative to prior studies, and shows massively parallel sequencing-based strategies can produce sufficient high quality sequence to reach support levels originally proposed for the phylogenetic bootstrap. Resampling simulations show that at least the entire plastome is necessary to fully resolve Pinus, particularly in rapidly radiating clades. Meta-analysis of 99 published infrageneric phylogenies shows that whole plastome analysis should provide similar gains across a range of plant genera. A disproportionate amount of phylogenetic information resides in two loci (ycf1, ycf2), highlighting their unusual evolutionary properties.
Conclusion: Plastome sequencing is now an efficient option for increasing phylogenetic resolution at lower taxonomic levels in plant phylogenetic and population genetic analyses. With continuing improvements in sequencing capacity, the strategies herein should revolutionize efforts requiring dense taxon and character sampling, such as phylogeographic analyses and species-level DNA barcoding.
Figures




References
-
- Philippe H, Frederic D, Henner B, Lartillot N. Phylogenomics. Annu Rev Ecol Evol Syst. 2005;36:541–542. doi: 10.1146/annurev.ecolsys.35.112202.130205. - DOI
-
- Jansen RK, Cai Z, Raubeson LA, Daniell H, Depamphilis CW, Leebens-Mack J, Muller KF, Guisinger-Bellian M, Haberle RC, Hansen AK. Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc Natl Acad Sci USA. 2007;104:19369. doi: 10.1073/pnas.0709121104. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

