Core promoter acetylation is not required for high transcription from the phosphoenolpyruvate carboxylase promoter in maize
- PMID: 19954517
- PMCID: PMC2793245
- DOI: 10.1186/1756-8935-2-17
Core promoter acetylation is not required for high transcription from the phosphoenolpyruvate carboxylase promoter in maize
Abstract
Background: Acetylation of promoter nucleosomes is tightly correlated and mechanistically linked to gene activity. However, transcription is not necessary for promoter acetylation. It seems, therefore, that external and endogenous stimuli control histone acetylation and by this contribute to gene regulation. Photosynthetic genes in plants are excellent models with which to study the connection between stimuli and chromatin modifications because these genes are strongly expressed and regulated by multiple stimuli that are easily manipulated. We have previously shown that acetylation of specific histone lysine residues on the photosynthetic phosphoenolpyruvate carboxylase (Pepc) promoter in maize is controlled by light and is independent of other stimuli or gene activity. Acetylation of upstream promoter regions responds to a set of other stimuli which include the nutrient availability of the plant. Here, we have extended these studies by analysing histone acetylation during the diurnal and circadian rhythm of the plant.
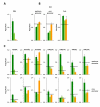
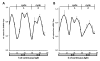
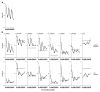
Results: We show that histone acetylation of individual lysine residues is removed from the core promoter before the end of the illumination period which is an indication that light is not the only factor influencing core promoter acetylation. Deacetylation is accompanied by a decrease in gene activity. Pharmacological inhibition of histone deacetylation is not sufficient to prevent transcriptional repression, indicating that deacetylation is not controlling diurnal gene regulation. Variation of the Pepc promoter activity during the day is controlled by the circadian oscillator as it is maintained under constant illumination for at least 3 days. During this period, light-induced changes in histone acetylation are completely removed from the core promoter, although the light stimulus is continuously applied. However, acetylation of most sites on upstream promoter elements follows the circadian rhythm.
Conclusion: Our results suggest a central role of upstream promoter acetylation in the quantitative regulation of gene expression in this model gene. Induced core promoter acetylation is dispensable for the highest gene expression in the diurnal and circadian rhythm.
Figures
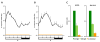

Similar articles
-
Illumination is necessary and sufficient to induce histone acetylation independent of transcriptional activity at the C4-specific phosphoenolpyruvate carboxylase promoter in maize.Plant Physiol. 2006 Jul;141(3):1078-88. doi: 10.1104/pp.106.080457. Epub 2006 May 5. Plant Physiol. 2006. PMID: 16679423 Free PMC article.
-
Developmental and environmental signals induce distinct histone acetylation profiles on distal and proximal promoter elements of the C4-Pepc gene in maize.Genetics. 2008 Aug;179(4):1891-901. doi: 10.1534/genetics.108.087411. Epub 2008 Aug 9. Genetics. 2008. PMID: 18689888 Free PMC article.
-
Circadian and light-induced transcription of clock gene Per1 depends on histone acetylation and deacetylation.Mol Cell Biol. 2004 Jul;24(14):6278-87. doi: 10.1128/MCB.24.14.6278-6287.2004. Mol Cell Biol. 2004. PMID: 15226430 Free PMC article.
-
Histone acetylation: facts and questions.Chromosoma. 1994 Dec;103(7):441-9. doi: 10.1007/BF00337382. Chromosoma. 1994. PMID: 7720410 Review.
-
Histone acetylation modifiers in the pathogenesis of malignant disease.Mol Med. 2000 Aug;6(8):623-44. Mol Med. 2000. PMID: 11055583 Free PMC article. Review.
Cited by
-
Interplay of light and temperature during the in planta modulation of C4 phosphoenolpyruvate carboxylase from the leaves of Amaranthus hypochondriacus L.: diurnal and seasonal effects manifested at molecular levels.J Exp Bot. 2011 Jan;62(3):1017-26. doi: 10.1093/jxb/erq333. Epub 2010 Nov 2. J Exp Bot. 2011. PMID: 21045006 Free PMC article.
-
A Common histone modification code on C4 genes in maize and its conservation in Sorghum and Setaria italica.Plant Physiol. 2013 May;162(1):456-69. doi: 10.1104/pp.113.216721. Epub 2013 Apr 5. Plant Physiol. 2013. PMID: 23564230 Free PMC article.
-
Photosynthetic Genes and Genes Associated with the C4 Trait in Maize Are Characterized by a Unique Class of Highly Regulated Histone Acetylation Peaks on Upstream Promoters.Plant Physiol. 2015 Aug;168(4):1378-88. doi: 10.1104/pp.15.00934. Epub 2015 Jun 25. Plant Physiol. 2015. PMID: 26111542 Free PMC article.
-
Major alterations in transcript profiles between C3-C4 and C4 photosynthesis of an amphibious species Eleocharis baldwinii.Plant Mol Biol. 2014 Sep;86(1-2):93-110. doi: 10.1007/s11103-014-0215-8. Epub 2014 Jul 10. Plant Mol Biol. 2014. PMID: 25008152
-
C4 Phosphoenolpyruvate Carboxylase: Evolution and transcriptional regulation.Genet Mol Biol. 2024 Mar 22;46(3 Suppl 1):e20230190. doi: 10.1590/1678-4685-GMB-2023-0190. eCollection 2024. Genet Mol Biol. 2024. PMID: 38517370 Free PMC article.
References
LinkOut - more resources
Full Text Sources

