Multiple invasions of Gypsy and Micropia retroelements in genus Zaprionus and melanogaster subgroup of the genus Drosophila
- PMID: 19954522
- PMCID: PMC2797524
- DOI: 10.1186/1471-2148-9-279
Multiple invasions of Gypsy and Micropia retroelements in genus Zaprionus and melanogaster subgroup of the genus Drosophila
Abstract
Background: The Zaprionus genus shares evolutionary features with the melanogaster subgroup, such as space and time of origin. Although little information about the transposable element content in the Zaprionus genus had been accumulated, some of their elements appear to be more closely related with those of the melanogaster subgroup, indicating that these two groups of species were involved in horizontal transfer events during their evolution. Among these elements, the Gypsy and the Micropia retroelements were chosen for screening in seven species of the two Zaprionus subgenera, Anaprionus and Zaprionus.
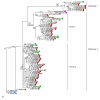
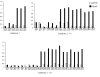
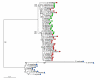
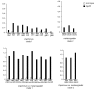
Results: Screening allowed the identification of diverse Gypsy and Micropia retroelements only in species of the Zaprionus subgenus, showing that they are transcriptionally active in the sampled species. The sequences of each retroelement were closely related to those of the melanogaster species subgroup, and the most parsimonious hypothesis would be that 15 horizontal transfer events shaped their evolution. The Gypsy retroelement of the melanogaster subgroup probably invaded the Zaprionus genomes about 11 MYA. In contrast, the Micropia retroelement may have been introduced into the Zaprionus subgenus and the melanogaster subgroup from an unknown donor more recently (~3 MYA).
Conclusion: Gypsy and Micropia of Zaprionus and melanogaster species share similar evolutionary patterns. The sharing of evolutionary, ecological and ethological features probably allowed these species to pass through a permissive period of transposable element invasion, explaining the proposed waves of horizontal transfers.
Figures

References
-
- Yassin A, Araripe LO, Capy P, Da Lage JL, Klaczko LB, Maisonhaute C, Ogereau D, David JR. Grafting the molecular phylogenetic tree with morphological branches to reconstruct the evolutionary history of the genus Zaprionus (Diptera: Drosophilidae) Mol Phylogenet Evol. 2008;47:903–15. doi: 10.1016/j.ympev.2008.01.036. - DOI - PubMed
-
- Russo CAM, Takezaki N, Nei M. Molecular phylogeny and divergence times of drosophilid species. Mol Biol Evol. 1995;12:391–404. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

