Sensitive plasma protein analysis by microparticle-based proximity ligation assays
- PMID: 19955079
- PMCID: PMC2830843
- DOI: 10.1074/mcp.M900248-MCP200
Sensitive plasma protein analysis by microparticle-based proximity ligation assays
Abstract
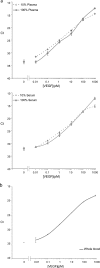
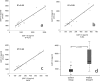
Detection of proteins released in the bloodstream from tissues damaged by disease can promote early detection of pathological conditions, differential diagnostics, and follow-up of therapy. Despite these prospects and a plethora of candidate biomarkers, efforts in recent years to establish new protein diagnostic assays have met with limited success. One important limiting factor has been the challenge of detecting proteins present at trace levels in complex bodily fluids. To achieve robust, sensitive, and specific detection, we have developed a microparticle-based solid-phase proximity ligation assay, dependent on simultaneous recognition of target proteins by three antibody molecules for added specificity. After capture on a microparticle, solid-phase pairs of proximity probes are added followed by washes, enabling detection and identification of rare protein molecules in blood while consuming small amounts of sample. We demonstrate that single polyclonal antibody preparations raised against target proteins of interest can be readily used to establish assays where detection depends on target recognition by three individual antibody molecules, recognizing separate epitopes. The assay was compared with state-of-the-art sandwich ELISAs for detection of vascular endothelial growth factor, interleukin-8 and interleukin-6, and it was found to be superior both with regard to dynamic range and minimal numbers of molecules detected. Furthermore, the assays exhibited excellent performance in undiluted plasma and serum as well as in whole blood, producing comparable results for nine different antigens. We thus show that solid-phase proximity ligation assay is suitable for validation of a variety of protein biomarkers over broad dynamic ranges in clinical samples.
Figures


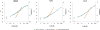

References
-
- Anderson N. L., Anderson N. G. ( 2002) The human plasma proteome: history, character, and diagnostic prospects. Mol. Cell. Proteomics 1, 845– 867 - PubMed
-
- Fredriksson S., Gullberg M., Jarvius J., Olsson C., Pietras K., Gústafsdóttir S. M., Ostman A., Landegren U. ( 2002) Protein detection using proximity-dependent DNA ligation assays. Nat. Biotechnol 20, 473– 477 - PubMed
-
- Gustafsdottir S. M., Nordengrahn A., Fredriksson S., Wallgren P., Rivera E., Schallmeiner E., Merza M., Landegren U. ( 2006) Detection of individual microbial pathogens by proximity ligation. Clin. Chem 52, 1152– 1160 - PubMed
-
- Söderberg O., Gullberg M., Jarvius M., Ridderstråle K., Leuchowius K. J., Jarvius J., Wester K., Hydbring P., Bahram F., Larsson L. G., Landegren U. ( 2006) Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat. Methods 3, 995– 1000 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

