Quantitative proteomics by metabolic labeling of model organisms
- PMID: 19955089
- PMCID: PMC2808257
- DOI: 10.1074/mcp.R900001-MCP200
Quantitative proteomics by metabolic labeling of model organisms
Abstract
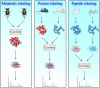
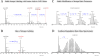
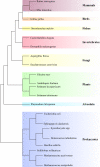
In the biological sciences, model organisms have been used for many decades and have enabled the gathering of a large proportion of our present day knowledge of basic biological processes and their derailments in disease. Although in many of these studies using model organisms, the focus has primarily been on genetics and genomics approaches, it is important that methods become available to extend this to the relevant protein level. Mass spectrometry-based proteomics is increasingly becoming the standard to comprehensively analyze proteomes. An important transition has been made recently by moving from charting static proteomes to monitoring their dynamics by simultaneously quantifying multiple proteins obtained from differently treated samples. Especially the labeling with stable isotopes has proved an effective means to accurately determine differential expression levels of proteins. Among these, metabolic incorporation of stable isotopes in vivo in whole organisms is one of the favored strategies. In this perspective, we will focus on methodologies to stable isotope label a variety of model organisms in vivo, ranging from relatively simple organisms such as bacteria and yeast to Caenorhabditis elegans, Drosophila, and Arabidopsis up to mammals such as rats and mice. We also summarize how this has opened up ways to investigate biological processes at the protein level in health and disease, revealing conservation and variation across the evolutionary tree of life.
Figures

References
-
- Bridges C. B. (1914) Direct proof through non-disjunction that the sex-linked genes of Drosophila are borne by the X-chromosome. Science 40, 107–109 - PubMed
-
- Lederberg J., Tatum E. L. (1946) Gene recombination in Escherichia coli. Nature 158, 558–558 - PubMed
-
- Hedges S. B. (2002) The origin and evolution of model organisms. Nat. Rev. Genet. 3, 838–849 - PubMed
-
- Hamann A., Brust D., Osiewacz H. D. (2008) Apoptosis pathways in fungal growth, development and ageing. Trends Microbiol. 16, 276–283 - PubMed
-
- Bowers K., Stevens T. H. (2005) Protein transport from the late Golgi to the vacuole in the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta 1744, 438–454 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

