Selection of DNA aptamers using atomic force microscopy
- PMID: 19955232
- PMCID: PMC2831320
- DOI: 10.1093/nar/gkp1101
Selection of DNA aptamers using atomic force microscopy
Abstract
Atomic force microscopy (AFM) can detect the adhesion or affinity force between a sample surface and cantilever, dynamically. This feature is useful as a method for the selection of aptamers that bind to their targets with very high affinity. Therefore, we propose the Systematic Evolution of Ligands by an EXponential enrichment (SELEX) method using AFM to obtain aptamers that have a strong affinity for target molecules. In this study, thrombin was chosen as the target molecule, and an 'AFM-SELEX' cycle was performed. As a result, selected cycles were completed with only three rounds, and many of the obtained aptamers had a higher affinity to thrombin than the conventional thrombin aptamer. Moreover, one type of obtained aptamer had a high affinity to thrombin as well as the anti-thrombin antibody. AFM-SELEX is, therefore, considered to be an available method for the selection of DNA aptamers that have a high affinity for their target molecules.
Figures

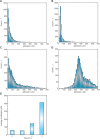
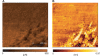

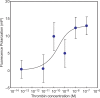
References
-
- Bass L, Cech R. Specific interaction between the self-splicing RNA of Tetrahymena and its guanosine substrate: implications for biological catalysis by RNA. Nature. 1984;308:820–826. - PubMed
-
- Gold L, Polisky B, Uhlenbeck O, Yarus M. Diversity of oligonucleotide functions. Annu. Rev. Biochem. 1995;64:763–797. - PubMed
-
- Osborne S, Ellington A. Nucleic acid selection and the challenge of combinatorial chemistry. Chem. Rev. 1997;97:349–370. - PubMed
-
- Wilson DS, Szostak Z. In vitro selection of functional nucleic acids. Annu. Rev. Biochem. 1999;68:647–661. - PubMed
-
- Misono TS, Kumar KR. Selection of RNA aptamers against human influenza virus hemagglutinin using surface plasmon resonance. Anal. Biochem. 2005;342:312–317. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

