An O-acetylserine(thiol)lyase homolog with L-cysteine desulfhydrase activity regulates cysteine homeostasis in Arabidopsis
- PMID: 19955263
- PMCID: PMC2815857
- DOI: 10.1104/pp.109.147975
An O-acetylserine(thiol)lyase homolog with L-cysteine desulfhydrase activity regulates cysteine homeostasis in Arabidopsis
Abstract
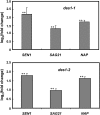
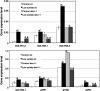
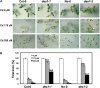
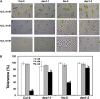
Cysteine (Cys) occupies a central position in plant metabolism due to its biochemical functions. Arabidopsis (Arabidopsis thaliana) cells contain different O-acetylserine(thiol)lyase (OASTL) enzymes that catalyze the biosynthesis of Cys. Because they are localized in the cytosol, plastids, and mitochondria, this results in multiple subcellular Cys pools. Much progress has been made on the most abundant OASTL enzymes; however, information on the less abundant OASTL-like proteins has been scarce. To unequivocally establish the enzymatic reaction catalyzed by the minor cytosolic OASTL isoform CS-LIKE (for Cys synthase-like; At5g28030), we expressed this enzyme in bacteria and characterized the purified recombinant protein. Our results demonstrate that CS-LIKE catalyzes the desulfuration of L-Cys to sulfide plus ammonia and pyruvate. Thus, CS-LIKE is a novel L-Cys desulfhydrase (EC 4.4.1.1), and we propose to designate it DES1. The impact and functionality of DES1 in Cys metabolism was revealed by the phenotype of the T-DNA insertion mutants des1-1 and des1-2. Mutation of the DES1 gene leads to premature leaf senescence, as demonstrated by the increased expression of senescence-associated genes and transcription factors. Also, the absence of DES1 significantly reduces the total Cys desulfuration activity in leaves, and there is a concomitant increase in the total Cys content. As a consequence, the expression levels of sulfur-responsive genes are deregulated, and the mutant plants show enhanced antioxidant defenses and tolerance to conditions that promote oxidative stress. Our results suggest that DES1 from Arabidopsis is an L-Cys desulfhydrase involved in maintaining Cys homeostasis, mainly at late developmental stages or under environmental perturbations.
Figures



References
-
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657 - PubMed
-
- Barroso C, Vega JM, Gotor C (1995) A new member of the cytosolic O-acetylserine(thiol)lyase gene family in Arabidopsis thaliana. FEBS Lett 363 1–5 - PubMed
-
- Berkowitz O, Wirtz M, Wolf A, Kuhlmann J, Hell R (2002) Use of biomolecular interaction analysis to elucidate the regulatory mechanism of the cysteine synthase complex from Arabidopsis thaliana. J Biol Chem 277 30629–30634 - PubMed
-
- Blaszczyk A, Brodzik R, Sirko A (1999) Increased resistance to oxidative stress in transgenic tobacco plants overexpressing bacterial serine acetyltransferase. Plant J 20 237–243 - PubMed
-
- Bogdanova N, Hell R (1997) Cysteine synthesis in plants: protein-protein interactions of serine acetyltransferase from Arabidopsis thaliana. Plant J 11 251–262 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

