Regional profiling for determination of genotype diversity of mastitis-specific Staphylococcus aureus lineage in Canada by use of clumping factor A, pulsed-field gel electrophoresis, and spa typing
- PMID: 19955267
- PMCID: PMC2815643
- DOI: 10.1128/JCM.01768-09
Regional profiling for determination of genotype diversity of mastitis-specific Staphylococcus aureus lineage in Canada by use of clumping factor A, pulsed-field gel electrophoresis, and spa typing
Abstract
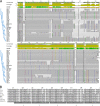
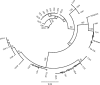
One of the major concerns in global public health and the dairy industry is the emergence of host-specific virulent Staphylococcus aureus strains. The high degree of stability of the species genome renders detection of genetic microvariations difficult. Thus, approaches for the rapid tracking of specialized lineages are urgently needed. We used clumping factor A (clfA) to profile 87 bovine mastitis isolates from four regions in Canada and compared the results to those obtained by pulsed-field gel electrophoresis (PFGE) and spa typing. Twenty-five pulsotypes were obtained by PFGE with an index of discrimination of 0.91. These were assigned to six PFGE lineage groups A to F and seven spa types, including two novel ones. Group A had 48.3% of the isolates and group D had 43.7% of the isolates, while only 8% of the isolates were variable. The results of antimicrobial susceptibility testing indicated that all isolates were sensitive to methicillin and the non-beta-lactam antibiotics, while three isolates were resistant to penicillin and one isolate was resistant to tetracycline. All isolates had the clfA gene and belonged to 20 clfA repeat types with an index of discrimination of 0.90. The dominant clfA types, types X, Q, C, and Z, formed 82% and 43% of PFGE groups A and D, respectively, and had copy numbers that varied only within a narrow range of between 46 and 52 copies, implying clonal selection. The rest were variable and region specific. Furthermore, the dominant groups contained subpopulations in different regions across Canada. Sequence information confirmed the relatedness obtained by the use of clfA repeat copy numbers and other methods and further revealed the occurrence of full-repeat deletions and conserved host-specific codon-triplet position biases at 18-bp units. Thus, concordant with the results of PFGE and spa typing, clfA typing proved useful for revealing the clonal nature of the mastitis isolate lineage and for the rapid profiling of subpopulations with comparable discriminatory powers.
Figures

References
-
- Asao, T., Y. Kumeda, T. Kawai, T. Shibata, H. Oda, K. Haruki, H. Nakazawa, and S. Kozaki. 2003. An extensive outbreak of staphylococcal food poisoning due to low-fat milk in Japan: estimation of enterotoxin A in the incriminated milk and powdered skim milk. Epidemiol. Infect. 130:33-40. - PMC - PubMed
-
- Atkins, K., J. Burman, E. Chamberlain, J. Cooper, B. Poutrel, S. Bagby, A. Jenkins, E. Feil, and J. van den Elsen. 2008. S. aureus IgG-binding proteins SpA and Sbi: host specificity and mechanisms of immune complex formation. Mol. Immunol. 45:1600-1611. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

