Coronavirus nucleocapsid protein facilitates template switching and is required for efficient transcription
- PMID: 19955314
- PMCID: PMC2812394
- DOI: 10.1128/JVI.02011-09
Coronavirus nucleocapsid protein facilitates template switching and is required for efficient transcription
Abstract
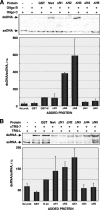
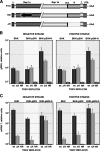
Purified nucleocapsid protein (N protein) from transmissible gastroenteritis virus (TGEV) enhanced hammerhead ribozyme self-cleavage and favored nucleic acid annealing, properties that define RNA chaperones, as previously reported. Several TGEV N-protein deletion mutants were expressed in Escherichia coli and purified, and their RNA binding ability and RNA chaperone activity were evaluated. The smallest N-protein domain analyzed with RNA chaperone activity, facilitating DNA and RNA annealing, contained the central unstructured region (amino acids 117 to 268). Interestingly, N protein and its deletion mutants with RNA chaperone activity enhanced template switching in a retrovirus-derived heterologous system, reinforcing the concept that TGEV N protein is an RNA chaperone that could be involved in template switching. This result is in agreement with the observation that in vivo, N protein is not necessary for TGEV replication, but it is required for efficient transcription.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

