Insulin-stimulated interaction with 14-3-3 promotes cytoplasmic localization of lipin-1 in adipocytes
- PMID: 19955570
- PMCID: PMC2823528
- DOI: 10.1074/jbc.M109.072488
Insulin-stimulated interaction with 14-3-3 promotes cytoplasmic localization of lipin-1 in adipocytes
Abstract
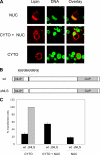
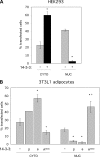
Lipin-1 is a bifunctional protein involved in lipid metabolism and adipogenesis. Lipin-1 plays a role in the biosynthesis of triacylglycerol through its phosphatidate phosphatase activity and also acts as a transcriptional co-activator of genes involved in oxidative metabolism. Lipin-1 resides in the cytoplasm and translocates to the endoplasmic reticulum membrane to catalyze the phosphatidate phosphatase reaction. It also possesses a nuclear localization signal, which is required for its translocation to the nucleus and may therefore be important for lipin-1 co-activator function. Thus, subcellular localization may be an important factor in the regulation of this protein. Here, we show that the nuclear localization signal alone is not sufficient for lipin-1 nuclear localization, and identify lipin-1 interaction with 14-3-3 as a determinant of its subcellular localization. We demonstrate that lipin-1 interacts with 14-3-3 proteins and that overexpression of 14-3-3 promotes the cytoplasmic localization of lipin-1 in 3T3-L1 adipocytes. The effect of 14-3-3 is mediated through a serine-rich domain in lipin-1. Functional mapping of the 14-3-3-interacting region within the serine-rich domain indicates redundancy and cooperativity among several sites, including five phosphorylated serine and threonine residues. Insulin stimulation of 3T3-L1 adipocytes results in increased lipin-1 phosphorylation, enhanced interaction with 14-3-3, and predominantly cytoplasmic localization. In summary, our studies suggest that insulin may modulate the cellular function of lipin-1 by regulating its subcellular localization through interactions with 14-3-3 proteins.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

