Exceptional structured noncoding RNAs revealed by bacterial metagenome analysis
- PMID: 19956260
- PMCID: PMC4140389
- DOI: 10.1038/nature08586
Exceptional structured noncoding RNAs revealed by bacterial metagenome analysis
Abstract
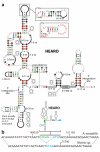
Estimates of the total number of bacterial species indicate that existing DNA sequence databases carry only a tiny fraction of the total amount of DNA sequence space represented by this division of life. Indeed, environmental DNA samples have been shown to encode many previously unknown classes of proteins and RNAs. Bioinformatics searches of genomic DNA from bacteria commonly identify new noncoding RNAs (ncRNAs) such as riboswitches. In rare instances, RNAs that exhibit more extensive sequence and structural conservation across a wide range of bacteria are encountered. Given that large structured RNAs are known to carry out complex biochemical functions such as protein synthesis and RNA processing reactions, identifying more RNAs of great size and intricate structure is likely to reveal additional biochemical functions that can be achieved by RNA. We applied an updated computational pipeline to discover ncRNAs that rival the known large ribozymes in size and structural complexity or that are among the most abundant RNAs in bacteria that encode them. These RNAs would have been difficult or impossible to detect without examining environmental DNA sequences, indicating that numerous RNAs with extraordinary size, structural complexity, or other exceptional characteristics remain to be discovered in unexplored sequence space.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

