Analysis of High-Throughput Flow Cytometry Data Using plateCore
- PMID: 19956418
- PMCID: PMC2777006
- DOI: 10.1155/2009/356141
Analysis of High-Throughput Flow Cytometry Data Using plateCore
Abstract
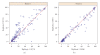
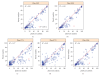
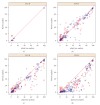
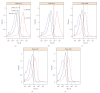
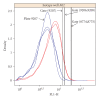
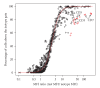
Flow cytometry (FCM) software packages from R/Bioconductor, such as flowCore and flowViz, serve as an open platform for development of new analysis tools and methods. We created plateCore, a new package that extends the functionality in these core packages to enable automated negative control-based gating and make the processing and analysis of plate-based data sets from high-throughput FCM screening experiments easier. plateCore was used to analyze data from a BD FACS CAP screening experiment where five Peripheral Blood Mononucleocyte Cell (PBMC) samples were assayed for 189 different human cell surface markers. This same data set was also manually analyzed by a cytometry expert using the FlowJo data analysis software package (TreeStar, USA). We show that the expression values for markers characterized using the automated approach in plateCore are in good agreement with those from FlowJo, and that using plateCore allows for more reproducible analyses of FCM screening data.
Figures

References
-
- Gasparetto M, Gentry T, Sebti S, et al. Identification of compounds that enhance the anti-lymphoma activity of rituximab using flow cytometric high-content screening. Journal of Immunological Methods. 2004;292(1-2):59–71. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

