Identification of the Schistosoma mansoni TNF-alpha receptor gene and the effect of human TNF-alpha on the parasite gene expression profile
- PMID: 19956564
- PMCID: PMC2779652
- DOI: 10.1371/journal.pntd.0000556
Identification of the Schistosoma mansoni TNF-alpha receptor gene and the effect of human TNF-alpha on the parasite gene expression profile
Abstract
Background: Schistosoma mansoni is the major causative agent of schistosomiasis. The parasite takes advantage of host signals to complete its development in the human body. Tumor necrosis factor-alpha (TNF-alpha) is a human cytokine involved in skin inflammatory responses, and although its effect on the adult parasite's metabolism and egg-laying process has been previously described, a comprehensive assessment of the TNF-alpha pathway and its downstream molecular effects is lacking.
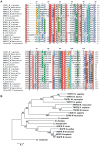
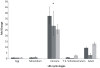
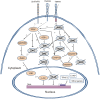
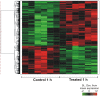
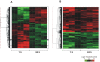
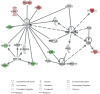
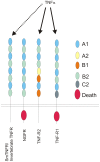
Methodology/principal findings: In the present work we describe a possible TNF-alpha receptor (TNFR) homolog gene in S. mansoni (SmTNFR). SmTNFR encodes a complete receptor sequence composed of 599 amino acids, and contains four cysteine-rich domains as described for TNFR members. Real-time RT-PCR experiments revealed that SmTNFR highest expression level is in cercariae, 3.5 (+/-0.7) times higher than in adult worms. Downstream members of the known human TNF-alpha pathway were identified by an in silico analysis, revealing a possible TNF-alpha signaling pathway in the parasite. In order to simulate parasite's exposure to human cytokine during penetration of the skin, schistosomula were exposed to human TNF-alpha just 3 h after cercariae-to-schistosomula in vitro transformation, and large-scale gene expression measurements were performed with microarrays. A total of 548 genes with significantly altered expression were detected, when compared to control parasites. In addition, treatment of adult worms with TNF-alpha caused a significantly altered expression of 1857 genes. Interestingly, the set of genes altered in adults is different from that of schistosomula, with 58 genes in common, representing 3% of altered genes in adults and 11% in 3 h-old early schistosomula.
Conclusions/significance: We describe the possible molecular elements and targets involved in human TNF-alpha effect on S. mansoni, highlighting the mechanism by which recently transformed schistosomula may sense and respond to this host mediator at the site of cercarial penetration into the skin.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
A quantitative proteomic analysis of the tegumental proteins from Schistosoma mansoni schistosomula reveals novel potential therapeutic targets.Int J Parasitol. 2015 Jul;45(8):505-16. doi: 10.1016/j.ijpara.2015.03.004. Epub 2015 Apr 21. Int J Parasitol. 2015. PMID: 25910674
-
Histamine signalling in Schistosoma mansoni: immunolocalisation and characterisation of a new histamine-responsive receptor (SmGPR-2).Int J Parasitol. 2010 Oct;40(12):1395-406. doi: 10.1016/j.ijpara.2010.04.006. Epub 2010 Apr 27. Int J Parasitol. 2010. PMID: 20430030
-
Molecular characterization of Schistosoma mansoni tegument annexins and comparative analysis of antibody responses following parasite infection.Mol Biochem Parasitol. 2019 Dec;234:111231. doi: 10.1016/j.molbiopara.2019.111231. Epub 2019 Oct 16. Mol Biochem Parasitol. 2019. PMID: 31628972
-
Receptors for growth and development of Schistosoma mansoni.J Helminthol. 2025 Feb 14;99:e29. doi: 10.1017/S0022149X24001020. J Helminthol. 2025. PMID: 39949117 Review.
-
Tissue-specific transcriptome analyses provide new insights into GPCR signalling in adult Schistosoma mansoni.PLoS Pathog. 2018 Jan 18;14(1):e1006718. doi: 10.1371/journal.ppat.1006718. eCollection 2018 Jan. PLoS Pathog. 2018. PMID: 29346437 Free PMC article. Review.
Cited by
-
Effects of proteasome inhibitor MG-132 on the parasite Schistosoma mansoni.PLoS One. 2017 Sep 12;12(9):e0184192. doi: 10.1371/journal.pone.0184192. eCollection 2017. PLoS One. 2017. PMID: 28898250 Free PMC article.
-
Effect of human TGF-β on the gene expression profile of Schistosoma mansoni adult worms.Mol Biochem Parasitol. 2012 Jun;183(2):132-9. doi: 10.1016/j.molbiopara.2012.02.008. Epub 2012 Feb 25. Mol Biochem Parasitol. 2012. PMID: 22387759 Free PMC article.
-
Curcumin Generates Oxidative Stress and Induces Apoptosis in Adult Schistosoma mansoni Worms.PLoS One. 2016 Nov 22;11(11):e0167135. doi: 10.1371/journal.pone.0167135. eCollection 2016. PLoS One. 2016. PMID: 27875592 Free PMC article.
-
The cell biology of schistosomes: a window on the evolution of the early metazoa.Protoplasma. 2012 Jul;249(3):503-18. doi: 10.1007/s00709-011-0326-x. Protoplasma. 2012. PMID: 21976269 Review.
-
Transcriptomic analysis of subarachnoid cysts of Taenia solium reveals mechanisms for uncontrolled proliferation and adaptations to the microenvironment.Sci Rep. 2024 May 23;14(1):11833. doi: 10.1038/s41598-024-61973-9. Sci Rep. 2024. PMID: 38782926 Free PMC article.
References
-
- WHO-Geneve. WHO Technical Report Series 912: prevention and control of schistosomiasis and soil-transmitted helminthiasis. Geneva: World Health Organization; 2002. - PubMed
-
- Pfeffer K. Biological functions of tumor necrosis factor cytokines and their receptors. Cytokine Growth Factor Rev. 2003;14:185–191. - PubMed
-
- He YX, Chen L, Ramaswamy K. Schistosoma mansoni, S. haematobium, and S. japonicum: early events associated with penetration and migration of schistosomula through human skin. Exp Parasitol. 2002;102:99–108. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

