SUMO modification regulates BLM and RAD51 interaction at damaged replication forks
- PMID: 19956565
- PMCID: PMC2779653
- DOI: 10.1371/journal.pbio.1000252
SUMO modification regulates BLM and RAD51 interaction at damaged replication forks
Abstract
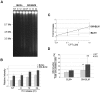
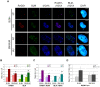
The gene mutated in Bloom's syndrome, BLM, is important in the repair of damaged replication forks, and it has both pro- and anti-recombinogenic roles in homologous recombination (HR). At damaged forks, BLM interacts with RAD51 recombinase, the essential enzyme in HR that catalyzes homology-dependent strand invasion. We have previously shown that defects in BLM modification by the small ubiquitin-related modifier (SUMO) cause increased gamma-H2AX foci. Because the increased gamma-H2AX could result from defective repair of spontaneous DNA damage, we hypothesized that SUMO modification regulates BLM's function in HR repair at damaged forks. To test this hypothesis, we treated cells that stably expressed a normal BLM (BLM+) or a SUMO-mutant BLM (SM-BLM) with hydroxyurea (HU) and examined the effects of stalled replication forks on RAD51 and its DNA repair functions. HU treatment generated excess gamma-H2AX in SM-BLM compared to BLM+ cells, consistent with a defect in replication-fork repair. SM-BLM cells accumulated increased numbers of DNA breaks and were hypersensitive to DNA damage. Importantly, HU treatment failed to induce sister-chromatid exchanges in SM-BLM cells compared to BLM+ cells, indicating a specific defect in HR repair and suggesting that RAD51 function could be compromised. Consistent with this hypothesis, RAD51 localization to HU-induced repair foci was impaired in SM-BLM cells. These data suggested that RAD51 might interact noncovalently with SUMO. We found that in vitro RAD51 interacts noncovalently with SUMO and that it interacts more efficiently with SUMO-modified BLM compared to unmodified BLM. These data suggest that SUMOylation controls the switch between BLM's pro- and anti-recombinogenic roles in HR. In the absence of BLM SUMOylation, BLM perturbs RAD51 localization at damaged replication forks and inhibits fork repair by HR. Conversely, BLM SUMOylation relieves its inhibitory effects on HR, and it promotes RAD51 function.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



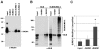

References
-
- Kowalczykowski S. C. Initiation of genetic recombination and recombination-dependent replication. Trends Biochem Sci. 2000;25:156–165. - PubMed
-
- Wilson D. M, 3rd, Thompson L. H. Molecular mechanisms of sister-chromatid exchange. Mutat Res. 2007;616:11–23. - PubMed
-
- Heartlein M. W, Tsuji H, Latt S. A. 5-Bromodeoxyuridine-dependent increase in sister chromatid exchange formation in Bloom's syndrome is associated with reduction in topoisomerase II activity. Exp Cell Res. 1987;169:245–254. - PubMed
-
- Palitti F, Tanzarella C, Degrassi F, De Salvia R, Fiore M. Enhancement of induced sister chromatid exchange and chromosomal aberrations by inhibitors of DNA repair processes. Toxicol Pathol. 1984;12:269–273. - PubMed
-
- Popescu N. C, Turnbull D, DiPaolo J. A. Sister chromatid exchange and chromosome aberration analysis with the use of several carcinogens and noncarcinogens. J Natl Cancer Inst. 1977;59:289–293. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

