The mycobacterial MsDps2 protein is a nucleoid-forming DNA binding protein regulated by sigma factors sigma and sigma
- PMID: 19956571
- PMCID: PMC2779847
- DOI: 10.1371/journal.pone.0008017
The mycobacterial MsDps2 protein is a nucleoid-forming DNA binding protein regulated by sigma factors sigma and sigma
Abstract
The Dps (DNA-binding protein from starved cells) proteins from Mycobacterium smegmatis MsDps1 and MsDps2 are both DNA-binding proteins with some differences. While MsDps1 has two oligomeric states, with one of them responsible for DNA binding, MsDps2 has only one DNA-binding oligomeric state. Both the proteins however, show iron-binding activity. The MsDps1 protein has been shown previously to be induced under conditions of starvation and osmotic stress and is regulated by the extra cellular sigma factors sigma(H) and sigma(F). We show here, that the second Dps homologue in M. smegmatis, namely MsDps2, is purified in a DNA-bound form and exhibits nucleoid-like structures under the atomic force microscope. It appears that the N-terminal sequence of Dps2 plays a role in nucleoid formation. MsDps2, unlike MsDps1, does not show elevated expression in nutritionally starved or stationary phase conditions; rather its promoter is recognized by RNA polymerase containing sigma(A) or sigma(B), under in vitro conditions. We propose that due to the nucleoid-condensing ability, the expression of MsDps2 is tightly regulated inside the cells.
Conflict of interest statement
Figures




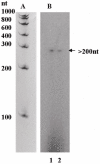

References
-
- Matin A, Auger EA, Blum PH, Schultz JE. Genetic basis of starvation survival in non differentiating bacteria. Annu Rev Microbiol. 1989;43:293–316. - PubMed
-
- Frenkiel-Krispin D, Minsky A. Nucleoid organization and the maintenance of DNA integrity in E. coli, B. subtilis and D. radiodurans. J Struct Biol. 2006;156:311–319. - PubMed
-
- Almirón M, Link AJ, Furlong D, Kolter R. A novel DNA-binding protein with regulatory and protective roles in starved Escherichia coli. Genes Dev. 1992;6:2646–2654. - PubMed
-
- Gupta S, Pandit SB, Srinivasan N, Chatterji D. Proteomics analysis of carbon-starved Mycobacterium smegmatis: induction of Dps-like protein. Protein Eng. 2002;15:503–512. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

