The E3 ubiquitin-ligase Bmi1/Ring1A controls the proteasomal degradation of Top2alpha cleavage complex - a potentially new drug target
- PMID: 19956605
- PMCID: PMC2779455
- DOI: 10.1371/journal.pone.0008104
The E3 ubiquitin-ligase Bmi1/Ring1A controls the proteasomal degradation of Top2alpha cleavage complex - a potentially new drug target
Abstract
Background: The topoisomerases Top1, Top2alpha and Top2beta are important molecular targets for antitumor drugs, which specifically poison Top1 or Top2 isomers. While it was previously demonstrated that poisoned Top1 and Top2beta are subject to proteasomal degradation, this phenomena was not demonstrated for Top2alpha.
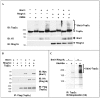
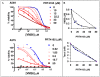
Methodology/principal findings: We show here that Top2alpha is subject to drug induced proteasomal degradation as well, although at a lower rate than Top2beta. Using an siRNA screen we identified Bmi1 and Ring1A as subunits of an E3 ubiquitin ligase involved in this process. We show that silencing of Bmi1 inhibits drug-induced Top2alpha degradation, increases the persistence of Top2alpha-DNA cleavage complex, and increases Top2 drug efficacy. The Bmi1/Ring1A ligase ubiquitinates Top2alpha in-vitro and cellular overexpression of Bmi1 increases drug induced Top2alpha ubiquitination. A small-molecular weight compound, identified in a screen for inhibitors of Bmi1/Ring1A ubiquitination activity, also prevents Top2alpha ubiquitination and drug-induced Top2alpha degradation. This ubiquitination inhibitor increases the efficacy of topoisomerase 2 poisons in a synergistic manner.
Conclusions/significance: The discovery that poisoned Top2alpha is undergoing proteasomal degradation combined with the involvement of Bmi1/Ring1A, allowed us to identify a small molecule that inhibits the degradation process. The Bmi1/Ring1A inhibitor sensitizes cells to Top2 drugs, suggesting that this type of drug combination will have a beneficial therapeutic outcome. As Bmi1 is also a known oncogene, elevated in numerous types of cancer, the identified Bmi1/Ring1A ubiquitin ligase inhibitors can also be potentially used to directly target the oncogenic properties of Bmi1.
Conflict of interest statement
Figures








References
-
- Holden JA. DNA topoisomerases as anticancer drug targets: from the laboratory to the clinic. Curr Med Chem Anticancer Agents. 2001;1:1–25. - PubMed
-
- Mao Y, Desai SD, Ting CY, Hwang J, Liu LF. 26 S proteasome-mediated degradation of topoisomerase II cleavable complexes. J Biol Chem. 2001;276:40652–40658. - PubMed
-
- Brown JM. The hypoxic cell: a target for selective cancer therapy–eighteenth Bruce F. Cain Memorial Award lecture. Cancer Res. 1999;59:5863–5870. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

