Epigenome microarray platform for proteome-wide dissection of chromatin-signaling networks
- PMID: 19956676
- PMCID: PMC2777412
- DOI: 10.1371/journal.pone.0006789
Epigenome microarray platform for proteome-wide dissection of chromatin-signaling networks
Abstract
Knowledge of protein domains that function as the biological effectors for diverse post-translational modifications of histones is critical for understanding how nuclear and epigenetic programs are established. Indeed, mutations of chromatin effector domains found within several proteins are associated with multiple human pathologies, including cancer and immunodeficiency syndromes. To date, relatively few effector domains have been identified in comparison to the number of modifications present on histone and non-histone proteins. Here we describe the generation and application of human modified peptide microarrays as a platform for high-throughput discovery of chromatin effectors and for epitope-specificity analysis of antibodies commonly utilized in chromatin research. Screening with a library containing a majority of the Royal Family domains present in the human proteome led to the discovery of TDRD7, JMJ2C, and MPP8 as three new modified histone-binding proteins. Thus, we propose that peptide microarray methodologies are a powerful new tool for elucidating molecular interactions at chromatin.
Conflict of interest statement
Figures
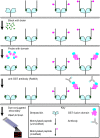


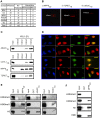
References
-
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403(6765):41–45. - PubMed
-
- Turner BM. Histone acetylation and an epigenetic code. Bioessays. 2000;22(9):836–845. - PubMed
-
- Bannister AJ, Kouzarides T. Histone methylation: Recognizing the methyl mark. Methods Enzymol. 2004;376:269–288. - PubMed
-
- Schreiber SL, Bernstein BE. Signaling network model of chromatin. Cell. 2002;111(6):771–778. - PubMed
-
- Schneider R, Bannister AJ, Myers FA, Thorne AW, Crane-Robinson C, et al. Histone H3 lysine 4 methylation patterns in higher eukaryotic genes. Nat Cell Biol. 2004;6(1):73–77. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

