Whole genome distribution and ethnic differentiation of copy number variation in Caucasian and Asian populations
- PMID: 19956714
- PMCID: PMC2776354
- DOI: 10.1371/journal.pone.0007958
Whole genome distribution and ethnic differentiation of copy number variation in Caucasian and Asian populations
Abstract
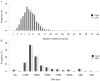
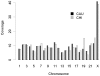
Although copy number variation (CNV) has recently received much attention as a form of structure variation within the human genome, knowledge is still inadequate on fundamental CNV characteristics such as occurrence rate, genomic distribution and ethnic differentiation. In the present study, we used the Affymetrix GeneChip(R) Mapping 500K Array to discover and characterize CNVs in the human genome and to study ethnic differences of CNVs between Caucasians and Asians. Three thousand and nineteen CNVs, including 2381 CNVs in autosomes and 638 CNVs in X chromosome, from 985 Caucasian and 692 Asian individuals were identified, with a mean length of 296 kb. Among these CNVs, 190 had frequencies greater than 1% in at least one ethnic group, and 109 showed significant ethnic differences in frequencies (p<0.01). After merging overlapping CNVs, 1135 copy number variation regions (CNVRs), covering approximately 439 Mb (14.3%) of the human genome, were obtained. Our findings of ethnic differentiation of CNVs, along with the newly constructed CNV genomic map, extend our knowledge on the structural variation in the human genome and may furnish a basis for understanding the genomic differentiation of complex traits across ethnic groups.
Conflict of interest statement
Figures


Similar articles
-
A genome-wide survey of copy number variations in Han Chinese residing in Taiwan.Genomics. 2009 Oct;94(4):241-6. doi: 10.1016/j.ygeno.2009.06.004. Epub 2009 Jun 25. Genomics. 2009. PMID: 19559783
-
Copy number variations (CNVs) identified in Korean individuals.BMC Genomics. 2008 Oct 18;9:492. doi: 10.1186/1471-2164-9-492. BMC Genomics. 2008. PMID: 18928558 Free PMC article.
-
Profiling of copy number variations (CNVs) in healthy individuals from three ethnic groups using a human genome 32 K BAC-clone-based array.Hum Mutat. 2008 Mar;29(3):398-408. doi: 10.1002/humu.20659. Hum Mutat. 2008. PMID: 18058796
-
Genomic Variation: Lessons Learned from Whole-Genome CNV Analysis.Curr Genet Med Rep. 2014 Jul 18;2(3):146-150. doi: 10.1007/s40142-014-0048-4. eCollection 2014. Curr Genet Med Rep. 2014. PMID: 25152847 Free PMC article. Review.
-
Gene copy number variation and common human disease.Clin Genet. 2010 Mar;77(3):201-13. doi: 10.1111/j.1399-0004.2009.01342.x. Epub 2009 Dec 10. Clin Genet. 2010. PMID: 20002459 Review.
Cited by
-
Compilation of copy number variants identified in phenotypically normal and parous Japanese women.J Hum Genet. 2014 Jun;59(6):326-31. doi: 10.1038/jhg.2014.27. Epub 2014 May 1. J Hum Genet. 2014. PMID: 24785687
-
Copy number variation of CCL3L1 among three major ethnic groups in Malaysia.BMC Genet. 2020 Jan 3;21(1):1. doi: 10.1186/s12863-019-0803-3. BMC Genet. 2020. PMID: 31900126 Free PMC article.
-
CNVIntegrate: the first multi-ethnic database for identifying copy number variations associated with cancer.Database (Oxford). 2021 Jul 14;2021:baab044. doi: 10.1093/database/baab044. Database (Oxford). 2021. PMID: 34259866 Free PMC article.
-
The Growing Importance of CNVs: New Insights for Detection and Clinical Interpretation.Front Genet. 2013 May 30;4:92. doi: 10.3389/fgene.2013.00092. eCollection 2013. Front Genet. 2013. PMID: 23750167 Free PMC article.
-
Copy number variation in the porcine genome inferred from a 60 k SNP BeadChip.BMC Genomics. 2010 Oct 22;11:593. doi: 10.1186/1471-2164-11-593. BMC Genomics. 2010. PMID: 20969757 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

