The fitness effects of random mutations in single-stranded DNA and RNA bacteriophages
- PMID: 19956760
- PMCID: PMC2776273
- DOI: 10.1371/journal.pgen.1000742
The fitness effects of random mutations in single-stranded DNA and RNA bacteriophages
Abstract
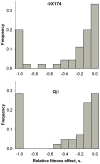
Mutational fitness effects can be measured with relatively high accuracy in viruses due to their small genome size, which facilitates full-length sequencing and genetic manipulation. Previous work has shown that animal and plant RNA viruses are very sensitive to mutation. Here, we characterize mutational fitness effects in single-stranded (ss) DNA and ssRNA bacterial viruses. First, we performed a mutation-accumulation experiment in which we subjected three ssDNA (PhiX174, G4, F1) and three ssRNA phages (Qbeta, MS2, and SP) to plaque-to-plaque transfers and chemical mutagenesis. Genome sequencing and growth assays indicated that the average fitness effect of the accumulated mutations was similar in the two groups. Second, we used site-directed mutagenesis to obtain 45 clones of PhiX174 and 42 clones of Qbeta carrying random single-nucleotide substitutions and assayed them for fitness. In PhiX174, 20% of such mutations were lethal, whereas viable ones reduced fitness by 13% on average. In Qbeta, these figures were 29% and 10%, respectively. It seems therefore that high mutational sensitivity is a general property of viruses with small genomes, including those infecting animals, plants, and bacteria. Mutational fitness effects are important for understanding processes of fitness decline, but also of neutral evolution and adaptation. As such, these findings can contribute to explain the evolution of ssDNA and ssRNA viruses.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- de Visser JA, Hermisson J, Wagner GP, Ancel ML, Bagheri-Chaichian H, et al. Perspective: Evolution and detection of genetic robustness. Evolution. 2003;57:1959–1972. - PubMed
-
- Ohta T. The nearly neutral theory of molecular evolution. Annu Rev Ecol Syst. 1992;23:263–286.
-
- Wagner A. New Jersey: Princeton University Press; 2005. Robustness and evolvability in living systems.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

