Linking human diseases to animal models using ontology-based phenotype annotation
- PMID: 19956802
- PMCID: PMC2774506
- DOI: 10.1371/journal.pbio.1000247
Linking human diseases to animal models using ontology-based phenotype annotation
Abstract
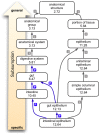
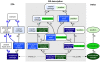
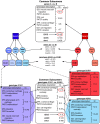
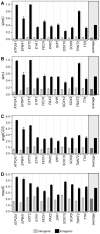
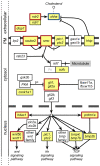
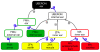
Scientists and clinicians who study genetic alterations and disease have traditionally described phenotypes in natural language. The considerable variation in these free-text descriptions has posed a hindrance to the important task of identifying candidate genes and models for human diseases and indicates the need for a computationally tractable method to mine data resources for mutant phenotypes. In this study, we tested the hypothesis that ontological annotation of disease phenotypes will facilitate the discovery of new genotype-phenotype relationships within and across species. To describe phenotypes using ontologies, we used an Entity-Quality (EQ) methodology, wherein the affected entity (E) and how it is affected (Q) are recorded using terms from a variety of ontologies. Using this EQ method, we annotated the phenotypes of 11 gene-linked human diseases described in Online Mendelian Inheritance in Man (OMIM). These human annotations were loaded into our Ontology-Based Database (OBD) along with other ontology-based phenotype descriptions of mutants from various model organism databases. Phenotypes recorded with this EQ method can be computationally compared based on the hierarchy of terms in the ontologies and the frequency of annotation. We utilized four similarity metrics to compare phenotypes and developed an ontology of homologous and analogous anatomical structures to compare phenotypes between species. Using these tools, we demonstrate that we can identify, through the similarity of the recorded phenotypes, other alleles of the same gene, other members of a signaling pathway, and orthologous genes and pathway members across species. We conclude that EQ-based annotation of phenotypes, in conjunction with a cross-species ontology, and a variety of similarity metrics can identify biologically meaningful similarities between genes by comparing phenotypes alone. This annotation and search method provides a novel and efficient means to identify gene candidates and animal models of human disease, which may shorten the lengthy path to identification and understanding of the genetic basis of human disease.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Collin G. B, Marshall J. D, Ikeda A, So W. V, Russell-Eggitt I, et al. Mutations in ALMS1 cause obesity, type 2 diabetes and neurosensory degeneration in Alstrom syndrome. Nat Genet. 2002;31:74–78. - PubMed
-
- Arsov T, Silva D. G, O'Bryan M. K, Sainsbury A, Lee N. J, et al. Fat aussie–a new Alstrom syndrome mouse showing a critical role for ALMS1 in obesity, diabetes, and spermatogenesis. Mol Endocrinol. 2006;20:1610–1622. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

