On primordial sense-antisense coding
- PMID: 19956936
- PMCID: PMC2853367
- DOI: 10.1007/s00239-009-9288-4
On primordial sense-antisense coding
Abstract
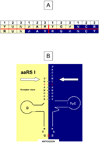
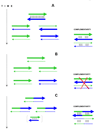
The genetic code is implemented by aminoacyl-tRNA synthetases (aaRS). These 20 enzymes are divided into two classes that, despite performing same functions, have nothing common in structure. The mystery of this striking partition of aaRSs might have been concealed in their sterically complementary modes of tRNA recognition that, as we have found recently, protect the tRNAs with complementary anticodons from confusion in translation. This finding implies that, in the beginning, life increased its coding repertoire by the pairs of complementary codons (rather than one-by-one) and used both complementary strands of genes as templates for translation. The class I and class II aaRSs may represent one of the most important examples of such primordial sense-antisense (SAS) coding (Rodin and Ohno, Orig Life Evol Biosph 25:565-589, 1995). In this report, we address the issue of SAS coding in a wider scope. We suggest a variety of advantages that such coding would have had in exploring a wider sequence space before translation became highly specific. In particular, we confirm that in Achlya klebsiana a single gene might have originally coded for an HSP70 chaperonin (class II aaRS homolog) and an NAD-specific GDH-like enzyme (class I aaRS homolog) via its sense and antisense strands. Thus, in contrast to the conclusions in Williams et al. (Mol Biol Evol 26:445-450, 2009), this could indeed be a "Rosetta stone" gene (Carter and Duax, Mol Cell 10:705-708, 2002) (eroded somewhat, though) for the SAS origin of the two aaRS classes.
Figures


References
-
- Branchiamore S, Riggs AD, Rodin SN. On the role of epigenetic silencing in evolution by gene duplication: Selection favors CpGs in coding regions of HOX genes – in preparation. 2009
-
- Carter CW, Jr, Duax WL. Did tRNA synthetase classes arise on opposite strands of the same gene? Mol. Cell. 2002;10:705–708. - PubMed
-
- Crick FHC. The origin of the genetic code. J. Mol. Biol. 1968;38:367–380. - PubMed
-
- Crick FHC, Brenner S, Klug A, Pieczenik G. A speculation on the origin of protein synthesis. Orig. Life. 1976;7:389–397. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

