DNA-binding residues and binding mode prediction with binding-mechanism concerned models
- PMID: 19958487
- PMCID: PMC2788376
- DOI: 10.1186/1471-2164-10-S3-S23
DNA-binding residues and binding mode prediction with binding-mechanism concerned models
Abstract
Background: Protein-DNA interactions are essential for fundamental biological activities including DNA transcription, replication, packaging, repair and rearrangement. Proteins interacting with DNA can be classified into two categories of binding mechanisms - sequence-specific and non-specific binding. Protein-DNA specific binding provides a mechanism to recognize correct nucleotide base pairs for sequence-specific identification. Protein-DNA non-specific binding shows sequence independent interaction for accelerated targeting by interacting with DNA backbone. Both sequence-specific and non-specific binding residues contribute to their roles for interaction.
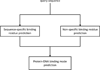
Results: The proposed framework has two stage predictors: DNA-binding residues prediction and binding mode prediction. In the first stage - DNA-binding residues prediction, the predictor for DNA specific binding residues achieves 96.45% accuracy with 50.14% sensitivity, 99.31% specificity, 81.70% precision, and 62.15% F-measure. The predictor for DNA non-specific binding residues achieves 89.14% accuracy with 53.06% sensitivity, 95.25% specificity, 65.47% precision, and 58.62% F-measure. While combining prediction results of sequence-specific and non-specific binding residues with OR operation, the predictor achieves 89.26% accuracy with 56.86% sensitivity, 95.63% specificity, 71.92% precision, and 63.51% F-measure. In the second stage, protein-DNA binding mode prediction achieves 75.83% accuracy while using support vector machine with multi-class prediction.
Conclusion: This article presents the design of a sequence based predictor aiming to identify sequence-specific and non-specific binding residues in a transcription factor with DNA binding-mechanism concerned. The protein-DNA binding mode prediction was introduced to help improve DNA-binding residues prediction. In addition, the results of this study will help with the design of binding-mechanism concerned predictors for other families of proteins interacting with DNA.
Figures
References
-
- Berman HM, Olson WK, Beveridge DL, Westbrook J, Gelbin A, Demeny T, Hsieh SH, Srinivasan AR, Schneider B. The nucleic acid database. A comprehensive relational database of three-dimensional structures of nucleic acids. Biophys J. 1992;63(3):751–759. doi: 10.1016/S0006-3495(92)81649-1. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

