Functional involvement of Tudor and dPRMT5 in the piRNA processing pathway in Drosophila germlines
- PMID: 19959991
- PMCID: PMC2797066
- DOI: 10.1038/emboj.2009.365
Functional involvement of Tudor and dPRMT5 in the piRNA processing pathway in Drosophila germlines
Abstract
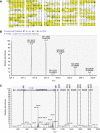
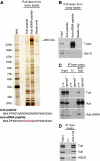
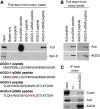
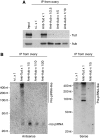
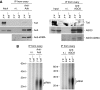
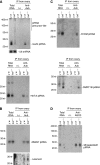
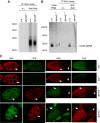
In Drosophila, the PIWI proteins, Aubergine (Aub), AGO3, and Piwi are expressed in germlines and function in silencing transposons by associating with PIWI-interacting RNAs (piRNAs). Recent studies show that PIWI proteins contain symmetric dimethyl-arginines (sDMAs) and that dPRMT5/Capsuleen/DART5 is the modifying enzyme. Here, we show that Tudor (Tud), one of Tud domain-containing proteins, associates with Aub and AGO3, specifically through their sDMA modifications and that these three proteins form heteromeric complexes. piRNA precursor-like molecules are detected in these complexes. The expression levels of Aub and AGO3, along with their degree of sDMA modification, were not changed by tud mutations. However, the population of transposon-derived piRNAs associated with Aub and AGO3 was altered by tud mutations, whereas the total amounts of small RNAs on Aub and AGO3 was increased. Loss of dprmt5 did not change the stability of Aub, but impaired its association with Tud and lowered piRNA association with Aub. Thus, in germline cells, piRNAs are quality-controlled by dPRMT5 that modifies PIWI proteins, in tight association with Tud.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
Similar articles
-
Arginine methylation of Piwi proteins catalysed by dPRMT5 is required for Ago3 and Aub stability.Nat Cell Biol. 2009 May;11(5):652-8. doi: 10.1038/ncb1872. Epub 2009 Apr 19. Nat Cell Biol. 2009. PMID: 19377467 Free PMC article.
-
Arginine methylation of Aubergine mediates Tudor binding and germ plasm localization.RNA. 2010 Jan;16(1):70-8. doi: 10.1261/rna.1869710. Epub 2009 Nov 19. RNA. 2010. PMID: 19926723 Free PMC article.
-
How does the royal family of Tudor rule the PIWI-interacting RNA pathway?Genes Dev. 2010 Apr 1;24(7):636-46. doi: 10.1101/gad.1899210. Genes Dev. 2010. PMID: 20360382 Free PMC article. Review.
-
Tudor-domain containing proteins act to make the piRNA pathways more robust in Drosophila.Fly (Austin). 2015;9(2):86-90. doi: 10.1080/19336934.2015.1128599. Fly (Austin). 2015. PMID: 26647059 Free PMC article.
-
piRNA-mediated silencing in Drosophila germlines.Semin Cell Dev Biol. 2010 Sep;21(7):754-9. doi: 10.1016/j.semcdb.2010.01.011. Epub 2010 Jan 18. Semin Cell Dev Biol. 2010. PMID: 20080197 Review.
Cited by
-
The genetic makeup of the Drosophila piRNA pathway.Mol Cell. 2013 Jun 6;50(5):762-77. doi: 10.1016/j.molcel.2013.04.031. Epub 2013 May 9. Mol Cell. 2013. PMID: 23665231 Free PMC article.
-
Mutations to the piRNA pathway component aubergine enhance meiotic drive of segregation distorter in Drosophila melanogaster.Genetics. 2013 Mar;193(3):771-84. doi: 10.1534/genetics.112.147561. Epub 2012 Dec 24. Genetics. 2013. PMID: 23267055 Free PMC article.
-
PRMT5 and the role of symmetrical dimethylarginine in chromatoid bodies of planarian stem cells.Development. 2012 Mar;139(6):1083-94. doi: 10.1242/dev.076182. Epub 2012 Feb 8. Development. 2012. PMID: 22318224 Free PMC article.
-
The lncRNA hsrω regulates arginine dimethylation of human FUS to cause its proteasomal degradation in Drosophila.J Cell Sci. 2019 Oct 23;132(20):jcs236836. doi: 10.1242/jcs.236836. J Cell Sci. 2019. PMID: 31519807 Free PMC article.
-
Tdrd6a Regulates the Aggregation of Buc into Functional Subcellular Compartments that Drive Germ Cell Specification.Dev Cell. 2018 Aug 6;46(3):285-301.e9. doi: 10.1016/j.devcel.2018.07.009. Dev Cell. 2018. PMID: 30086300 Free PMC article.
References
-
- Anne J, Mechler BM (2005) Valois, a component of the nuage and pole plasm, is involved in assembly of these structures, and binds to Tudor and the methyltransferase Capsuléen. Development 132: 2167–2177 - PubMed
-
- Anne J, Ollo R, Ephrussi A, Mechler BM (2007) Arginine methyltransferase Capsuleen is essential for methylation of spliceosomal Sm proteins and germ cell formation in Drosophila. Development 134: 137–146 - PubMed
-
- Aravin AA, Hannon GJ, Brennecke J (2007) The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science 318: 761–764 - PubMed
-
- Arkov AL, Wang JY, Ramos A, Lehmann R (2006) The role of Tudor domains in germline development and polar granule architecture. Development 133: 4053–4062 - PubMed
-
- Bardsley A, McDonald K, Boswell RE (1993) Distribution of tudor protein in the Drosophila embryo suggests separation of functions based on site of localization. Development 119: 207–219 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

