Conformational changes in the nicotinic acetylcholine receptor during gating and desensitization
- PMID: 19961216
- PMCID: PMC2818735
- DOI: 10.1021/bi901550p
Conformational changes in the nicotinic acetylcholine receptor during gating and desensitization
Abstract
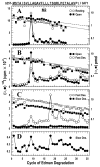
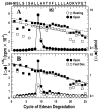
The nicotinic acetylcholine receptor (nAChR) is a member of the important Cys loop ligand-gated ion channel superfamily that modulates neuronal excitability. After they respond to their agonists, their actions are terminated either by removal of ligand or by fast and slow desensitization, processes that play an important role in modulating the duration of conducting states and hence of integrated neuronal behavior. We monitored structural changes occurring during fast and slow desensitization in the transmembrane domain of the Torpedo nAChR using time-resolved photolabeling with the hydrophobic probe 3-(trifluoromethyl)-3-(m-iodophenyl)diazirine (TID). After channel opening, TID photolabels a residue on the delta-subunit's M2-M3 loop and a cluster of four residues on deltaM1 and deltaM2, defining an open state pocket [Arevalo, E., et al. (2005) J. Biol. Chem. 280, 13631-13640]. We now find that photolabeling of this pocket persists during the transition to the fast desensitized state, the extent of photoincorporation decreasing only with the transition to the slow desensitized state. In contrast, the extent of photoincorporation in the channel lumen at the conserved 9'-leucines on the second transmembrane helix (M2-9') decreased successively during the resting to open and open to fast desensitized state transitions, implying that the local conformation is different in each state, a conclusion consistent with the hypothesis that there are separate gates for channel opening and desensitization. Thus, although during fast desensitization there is a conformation change in the channel lumen at the level of M2-9', there is none in the regions of the delta-subunit's M2-M3 loop and the interior of its M1-M4 helix bundle until slow desensitization occurs.
Figures



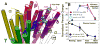
Similar articles
-
Gating-enhanced accessibility of hydrophobic sites within the transmembrane region of the nicotinic acetylcholine receptor's {delta}-subunit. A time-resolved photolabeling study.J Biol Chem. 2005 Apr 8;280(14):13631-40. doi: 10.1074/jbc.M413911200. Epub 2005 Jan 21. J Biol Chem. 2005. PMID: 15664985
-
Probing the structure of the affinity-purified and lipid-reconstituted torpedo nicotinic acetylcholine receptor.Biochemistry. 2008 Dec 2;47(48):12787-94. doi: 10.1021/bi801476j. Biochemistry. 2008. PMID: 18991407 Free PMC article.
-
[3H]Benzophenone photolabeling identifies state-dependent changes in nicotinic acetylcholine receptor structure.Biochemistry. 2007 Sep 11;46(36):10296-307. doi: 10.1021/bi7008163. Epub 2007 Aug 9. Biochemistry. 2007. PMID: 17685589
-
Assembly of nicotinic and other Cys-loop receptors.J Neurochem. 2011 Mar;116(5):734-41. doi: 10.1111/j.1471-4159.2010.07060.x. Epub 2011 Jan 13. J Neurochem. 2011. PMID: 21214570 Review.
-
Gating of nicotinic ACh receptors; new insights into structural transitions triggered by agonist binding that induce channel opening.J Physiol. 2007 Nov 1;584(Pt 3):727-33. doi: 10.1113/jphysiol.2007.142554. Epub 2007 Sep 6. J Physiol. 2007. PMID: 17823204 Free PMC article. Review.
Cited by
-
Desformylflustrabromine (dFBr) and [3H]dFBr-Labeled Binding Sites in a Nicotinic Acetylcholine Receptor.Mol Pharmacol. 2015 Jul;88(1):1-11. doi: 10.1124/mol.115.098913. Epub 2015 Apr 13. Mol Pharmacol. 2015. PMID: 25870334 Free PMC article.
-
A locally closed conformation of a bacterial pentameric proton-gated ion channel.Nat Struct Mol Biol. 2012 May 13;19(6):642-9. doi: 10.1038/nsmb.2307. Nat Struct Mol Biol. 2012. PMID: 22580559
-
Photoaffinity labeling the propofol binding site in GLIC.Biochemistry. 2014 Jan 14;53(1):135-42. doi: 10.1021/bi401492k. Epub 2013 Dec 30. Biochemistry. 2014. PMID: 24341978 Free PMC article.
-
Activation and desensitization induce distinct conformational changes at the extracellular-transmembrane domain interface of the glycine receptor.J Biol Chem. 2011 Nov 4;286(44):38814-38824. doi: 10.1074/jbc.M111.273631. Epub 2011 Sep 14. J Biol Chem. 2011. PMID: 21917927 Free PMC article.
-
Enantiomeric barbiturates bind distinct inter- and intrasubunit binding sites in a nicotinic acetylcholine receptor (nAChR).J Biol Chem. 2017 Oct 20;292(42):17258-17271. doi: 10.1074/jbc.M117.808592. Epub 2017 Sep 6. J Biol Chem. 2017. PMID: 28878016 Free PMC article.
References
-
- Cederholm JM, Schofield PR, Lewis TM. Gating mechanisms in Cys-loop receptors. Eur Biophys J. 2009 in press. - PubMed
-
- Sine SM, Engel AG. Recent advances in Cys-loop receptor structure and function. Nature. 2006;440:448–455. - PubMed
-
- Giniatullin R, Nistri A, Yakel JL. Desensitization of nicotinic ACh receptors: shaping cholinergic signaling. Trends Neurosci. 2005;28:371–378. - PubMed
-
- Unwin N. Refined structure of the nicotinic acetylcholine receptor at 4A resolution. Journal of molecular biology. 2005;346:967–989. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

